You are browsing environment: FUNGIDB
CAZyme Information: KNZ44280.1
Basic Information
help
| Species |
Puccinia sorghi
|
| Lineage |
Basidiomycota; Pucciniomycetes; ; Pucciniaceae; Puccinia; Puccinia sorghi
|
| CAZyme ID |
KNZ44280.1
|
| CAZy Family |
GT90 |
| CAZyme Description |
GH26 domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A0L6U920]
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 482 |
|
53060.04 |
5.1874 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PsorghiRO10H11247 |
21769 |
N/A |
691 |
21078
|
|
| Gene Location |
No EC number prediction in KNZ44280.1.
KNZ44280.1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 3.43e-111 |
55 |
405 |
44 |
409 |
| 3.02e-110 |
50 |
393 |
29 |
385 |
| 1.12e-63 |
186 |
455 |
1 |
267 |
| 4.73e-36 |
50 |
379 |
59 |
391 |
| 1.84e-35 |
50 |
379 |
63 |
395 |
KNZ44280.1 has no PDB hit.
KNZ44280.1 has no Swissprot hit.
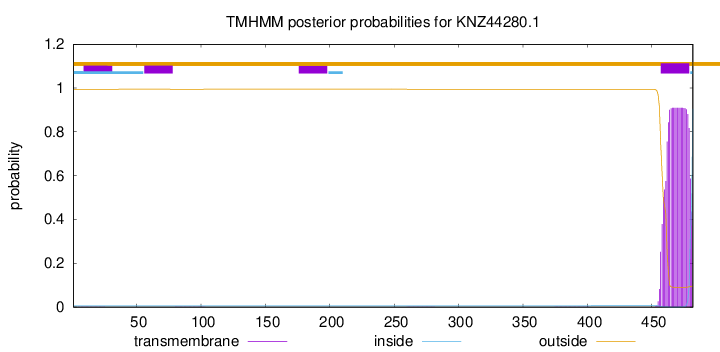
This protein is predicted as SP
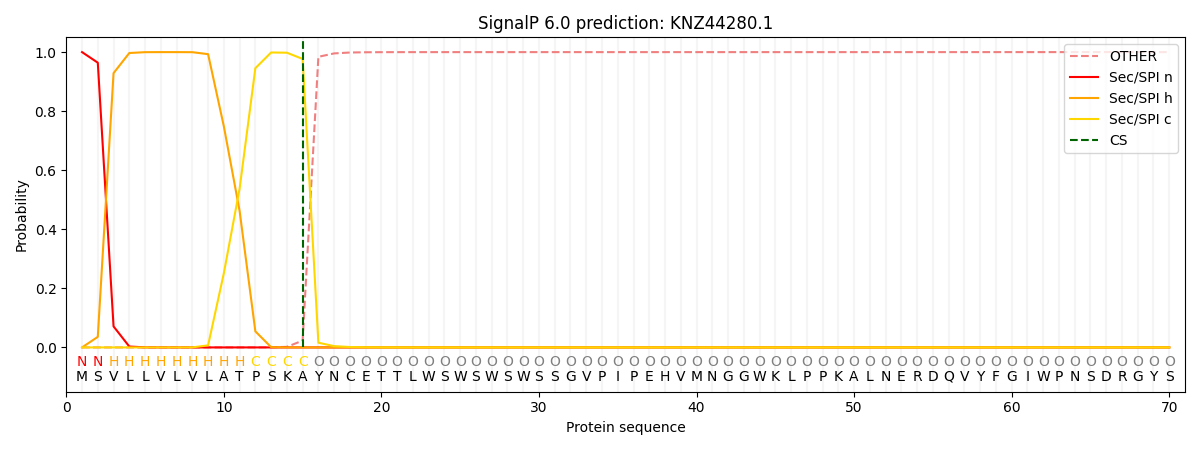
| Other |
SP_Sec_SPI |
CS Position |
| 0.000269 |
0.999688 |
CS pos: 15-16. Pr: 0.9768 |