You are browsing environment: FUNGIDB
CAZyme Information: KLU92743.1
You are here: Home > Sequence: KLU92743.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Magnaporthiopsis poae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Magnaporthiopsis; Magnaporthiopsis poae | |||||||||||
| CAZyme ID | KLU92743.1 | |||||||||||
| CAZy Family | GT41 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 227 | 442 | 2.8e-35 | 0.920704845814978 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225256 | Fes | 7.48e-16 | 231 | 452 | 87 | 299 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| 395613 | Esterase | 5.13e-07 | 228 | 378 | 10 | 150 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| 199889 | E_set_AMPKbeta_like_N | 2.31e-05 | 106 | 166 | 25 | 72 | N-terminal Early set domain, a glycogen binding domain, associated with the catalytic domain of AMP-activated protein kinase beta subunit. E or "early" set domains are associated with the catalytic domain of AMP-activated protein kinase beta subunit glycogen binding domain at the N-terminal end. AMPK is a metabolic stress sensing protein that senses AMP/ATP and has recently been found to act as a glycogen sensor as well. The protein functions as an alpha-beta-gamma heterotrimer. This N-terminal domain is the glycogen binding domain of the beta subunit. This domain is also a member of the CBM48 (Carbohydrate Binding Module 48) family whose members include pullulanase, maltooligosyl trehalose synthase, starch branching enzyme, glycogen branching enzyme, glycogen debranching enzyme, and isoamylase. |
| 225375 | YbbA | 5.10e-05 | 230 | 372 | 27 | 171 | Predicted hydrolase of the alpha/beta superfamily [General function prediction only]. |
| 406865 | AMPK1_CBM | 0.006 | 123 | 159 | 39 | 64 | Glycogen recognition site of AMP-activated protein kinase. AMPK1_CBM is a family found in close association with AMPKBI pfam04739. The surface of AMPK1_CBM reveals a carbohydrate-binding pocket. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.86e-47 | 26 | 459 | 50 | 461 | |
| 2.03e-28 | 230 | 451 | 67 | 284 | |
| 7.50e-28 | 106 | 448 | 329 | 640 | |
| 2.65e-27 | 230 | 451 | 67 | 284 | |
| 6.73e-26 | 106 | 443 | 332 | 647 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.95e-26 | 230 | 445 | 135 | 340 | Crystal structure of the N-terminal carbohydrate binding module family 48 and ferulic acid esterase from the multi-enzyme CE1-GH62-GH10 [uncultured bacterium] |
|
| 6.05e-26 | 230 | 445 | 135 | 340 | Crystal structure of the N-terminal carbohydrate binding module family 48 and ferulic acid esterase from the multi-enzyme CE1-GH62-GH10 [uncultured bacterium] |
|
| 7.00e-25 | 118 | 443 | 67 | 380 | Crystal structure of the N-terminal carbohydrate binding module family 48 and ferulic acid esterase from the multi-enzyme CE1-GH62-GH10 [uncultured bacterium],6RZN_B Crystal structure of the N-terminal carbohydrate binding module family 48 and ferulic acid esterase from the multi-enzyme CE1-GH62-GH10 [uncultured bacterium] |
|
| 1.12e-23 | 205 | 458 | 29 | 268 | Chain A, Endo-1,4-beta-xylanase Z [Acetivibrio thermocellus] |
|
| 2.87e-23 | 205 | 458 | 29 | 268 | Chain A, PROTEIN (ENDO-1,4-BETA-XYLANASE Z) [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.50e-21 | 106 | 443 | 356 | 664 | Carbohydrate acetyl esterase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe1-6A PE=1 SV=1 |
|
| 1.79e-21 | 205 | 458 | 48 | 287 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
|
| 1.16e-15 | 85 | 440 | 405 | 717 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
|
| 3.80e-12 | 170 | 371 | 801 | 979 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
|
| 3.82e-11 | 230 | 403 | 161 | 333 | Uncharacterized protein YieL OS=Escherichia coli (strain K12) OX=83333 GN=yieL PE=4 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
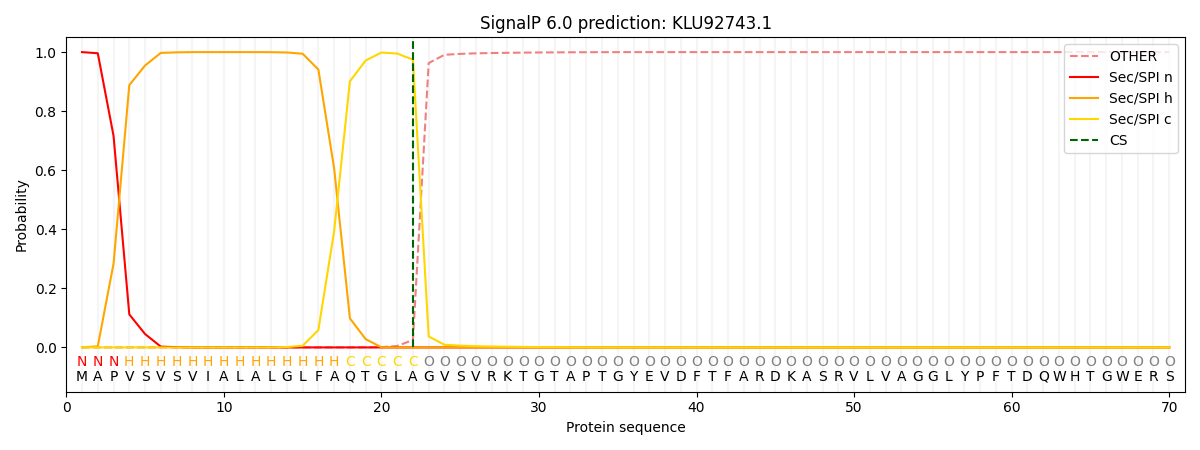
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000303 | 0.999667 | CS pos: 22-23. Pr: 0.9747 |
