You are browsing environment: FUNGIDB
CAZyme Information: KLU92247.1
You are here: Home > Sequence: KLU92247.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
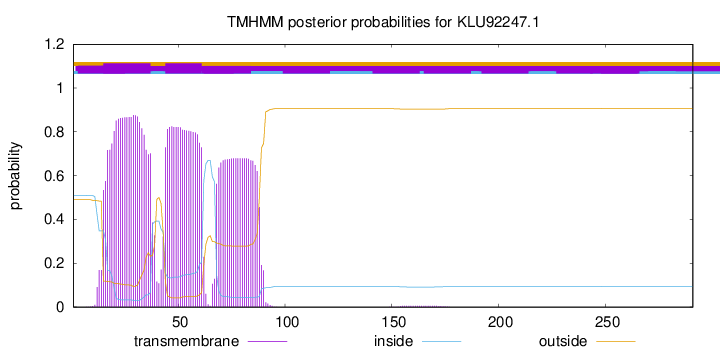
TMHMM annotations
Basic Information help
| Species | Magnaporthiopsis poae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Magnaporthiopsis; Magnaporthiopsis poae | |||||||||||
| CAZyme ID | KLU92247.1 | |||||||||||
| CAZy Family | GT22 | |||||||||||
| CAZyme Description | cutinase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE5 | 115 | 287 | 5.3e-39 | 0.9788359788359788 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395860 | Cutinase | 2.83e-42 | 115 | 286 | 1 | 169 | Cutinase. |
| 380565 | PBP1_ABC_LIVBP-like | 0.007 | 131 | 215 | 116 | 211 | type 1 periplasmic ligand-binding domain of ABC (Atpase Binding Cassette)-type active transport systems involved in the transport of all three branched chain aliphatic amino acids (leucine, isoleucine and valine). This subgroup includes the type 1 periplasmic ligand-binding domain of ABC (Atpase Binding Cassette)-type active transport systems that are involved in the transport of all three branched chain aliphatic amino acids (leucine, isoleucine and valine). This subgroup also includes a leucine-specific binding protein (or LivK), which is very similar in sequence and structure to leucine-isoleucine-valine binding protein (LIVBP). ABC-type active transport systems are transmembrane proteins that function in the transport of diverse sets of substrates across extra- and intracellular membranes, including carbohydrates, amino acids, inorganic ions, dipeptides and oligopeptides, metabolic products, lipids and sterols, and heme, to name a few. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.25e-81 | 66 | 290 | 1 | 214 | |
| 4.88e-57 | 72 | 290 | 6 | 220 | |
| 3.60e-55 | 98 | 275 | 20 | 197 | |
| 1.02e-53 | 78 | 287 | 12 | 221 | |
| 9.40e-52 | 66 | 287 | 1 | 216 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.15e-49 | 95 | 286 | 2 | 194 | Chain A, cutinase [Malbranchea cinnamomea] |
|
| 7.28e-38 | 96 | 275 | 8 | 191 | Chain A, CUTINASE [Fusarium vanettenii],1CUW_B Chain B, CUTINASE [Fusarium vanettenii] |
|
| 1.03e-37 | 96 | 275 | 8 | 191 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 1.44e-37 | 96 | 275 | 8 | 191 | Chain A, CUTINASE [Fusarium vanettenii] |
|
| 2.85e-37 | 96 | 275 | 8 | 191 | Chain A, CUTINASE [Fusarium vanettenii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.00e-82 | 66 | 290 | 1 | 214 | Cutinase CUT2 OS=Magnaporthe oryzae (strain 70-15 / ATCC MYA-4617 / FGSC 8958) OX=242507 GN=CUT2 PE=1 SV=1 |
|
| 1.26e-47 | 66 | 286 | 1 | 207 | Probable cutinase 1 OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=ACLA_081180 PE=3 SV=1 |
|
| 1.77e-47 | 83 | 290 | 4 | 211 | Probable cutinase 1 OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=AFUB_025250 PE=3 SV=1 |
|
| 1.77e-47 | 83 | 290 | 4 | 211 | Probable cutinase 1 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=AFUA_2G09380 PE=3 SV=1 |
|
| 1.11e-45 | 83 | 290 | 4 | 211 | Probable cutinase 1 OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=NFIA_084890 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999804 | 0.000179 |