You are browsing environment: FUNGIDB
CAZyme Information: KLU87244.1
You are here: Home > Sequence: KLU87244.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Magnaporthiopsis poae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Magnaporthiopsis; Magnaporthiopsis poae | |||||||||||
| CAZyme ID | KLU87244.1 | |||||||||||
| CAZy Family | GH27 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH55 | 156 | 910 | 3.4e-191 | 0.9743243243243244 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 176558 | PI-PLCXDc_like_2 | 5.00e-99 | 1102 | 1404 | 1 | 300 | Catalytic domain of uncharacterized hypothetical proteins similar to eukaryotic phosphatidylinositol-specific phospholipase C, X domain containing proteins. This subfamily corresponds to the catalytic domain present in a group of uncharacterized hypothetical proteins found in bacteria and fungi, which are similar to eukaryotic phosphatidylinositol-specific phospholipase C, X domain containing proteins (PI-PLCXD). The typical eukaryotic phosphoinositide-specific phospholipase C (PI-PLC, EC 3.1.4.11) has a multidomain organization that consists of a PLC catalytic core domain, and various regulatory domains. The catalytic core domain is assembled from two highly conserved X- and Y-regions split by a divergent linker sequence. In contrast, eukaryotic PI-PLCXDs contain a single TIM-barrel type catalytic domain, X domain, and are more closely related to bacterial PI-PLCs, which participate in Ca2+-independent PI metabolism, hydrolyzing the membrane lipid phosphatidylinositol (PI) to produce phosphorylated myo-inositol and diacylglycerol (DAG). Although the biological function of eukaryotic PI-PLCXDs still remains unclear, it may distinct from that of typical eukaryotic PI-PLCs. |
| 403800 | Pectate_lyase_3 | 7.88e-57 | 183 | 414 | 1 | 213 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| 176529 | PI-PLCXDc_like | 5.96e-49 | 1103 | 1381 | 2 | 277 | Catalytic domain of phosphatidylinositol-specific phospholipase C X domain containing and similar proteins. This family corresponds to the catalytic domain present in phosphatidylinositol-specific phospholipase C X domain containing proteins (PI-PLCXD) which are bacterial phosphatidylinositol-specific phospholipase C (PI-PLC, EC 4.6.1.13) sequence homologs mainly found in eukaryota. The typical eukaryotic phosphoinositide-specific phospholipase C (PI-PLC, EC 3.1.4.11) have a multidomain organization that consists of a PLC catalytic core domain, and various regulatory domains. The catalytic core domain is assembled from two highly conserved X- and Y-regions split by a divergent linker sequence. In contrast, eukaryotic PI-PLCXDs and their bacterial homologs contain a single TIM-barrel type catalytic domain, X domain, which is more closely related to that of bacterial PI-PLCs. Although the biological function of eukaryotic PI-PLCXDs still remains unclear, it may be distinct from that of typical eukaryotic PI-PLCs. |
| 176500 | PI-PLCc_bacteria_like | 1.55e-35 | 1103 | 1381 | 2 | 260 | Catalytic domain of bacterial phosphatidylinositol-specific phospholipase C and similar proteins. This subfamily corresponds to the catalytic domain present in bacterial phosphatidylinositol-specific phospholipase C (PI-PLC, EC 4.6.1.13) and their sequence homologs found in eukaryota. Bacterial PI-PLCs participate in Ca2+-independent PI metabolism, hydrolyzing the membrane lipid phosphatidylinositol (PI) to produce phosphorylated myo-inositol and diacylglycerol (DAG). Although their precise physiological function remains unclear, bacterial PI-PLCs may function as virulence factors in some pathogenic bacteria. Bacterial PI-PLCs contain a single TIM-barrel type catalytic domain. Its catalytic mechanism is based on general base and acid catalysis utilizing two well conserved histidines, and consists of two steps, a phosphotransfer and a phosphodiesterase reaction. Eukaryotic homologs in this family are named as phosphatidylinositol-specific phospholipase C X domain containing proteins (PI-PLCXD). They are distinct from the typical eukaryotic phosphoinositide-specific phospholipases C (PI-PLC, EC 3.1.4.11), which have a multidomain organization that consists of a PLC catalytic core domain, and various regulatory domains. The catalytic core domain is assembled from two highly conserved X- and Y-regions split by a divergent linker sequence. In contrast, eukaryotic PI-PLCXDs contain a single TIM-barrel type catalytic domain, X domain, which is closely related to that of bacterial PI-PLCs. Although the biological function of eukaryotic PI-PLCXDs still remains unclear, it may be distinct from that of typical eukaryotic PI-PLCs. This family also includes a distinctly different type of eukaryotic PLC, glycosylphosphatidylinositol-specific phospholipase C (GPI-PLC), an integral membrane protein characterized in the protozoan parasite Trypanosoma brucei. T. brucei GPI-PLC hydrolyzes the GPI-anchor on the variant specific glycoprotein (VSG), releasing dimyristyl glycerol (DMG), which may facilitate the evasion of the protozoan to the host's immune system. It does not require Ca2+ for its activity and is more closely related to bacterial PI-PLCs, but not mammalian PI-PLCs. |
| 176555 | PI-PLCXD1c | 9.34e-09 | 1110 | 1381 | 10 | 279 | Catalytic domain of phosphatidylinositol-specific phospholipase C, X domain containing 1. This subfamily corresponds to the catalytic domain present in a group of phosphatidylinositol-specific phospholipase C X domain containing 1 (PI-PLCXD1), 2 (PI-PLCXD2) and 3 (PI-PLCXD3), which are bacterial phosphatidylinositol-specific phospholipase C (PI-PLC, EC 4.6.1.13) sequence homologs found in vertebrates. The typical eukaryotic phosphoinositide-specific phospholipase C (PI-PLC, EC 3.1.4.11) has a multidomain organization that consists of a PLC catalytic core domain, and various regulatory domains. The catalytic core domain is assembled from two highly conserved X- and Y-regions split by a divergent linker sequence. In contrast, members in this group contain a single TIM-barrel type catalytic domain, X domain, and are more closely related to bacterial PI-PLCs, which participate in Ca2+-independent PI metabolism, hydrolyzing the membrane lipid phosphatidylinositol (PI) to produce phosphorylated myo-inositol and diacylglycerol (DAG). Although the biological function of eukaryotic PI-PLCXDs still remains unclear, it may distinct from that of typical eukaryotic PI-PLCs. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 34 | 1420 | 35 | 1431 | |
| 0.0 | 18 | 1419 | 29 | 1445 | |
| 8.61e-298 | 337 | 1419 | 9 | 1106 | |
| 1.70e-188 | 18 | 949 | 58 | 974 | |
| 1.10e-187 | 47 | 927 | 441 | 1297 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.67e-120 | 163 | 906 | 4 | 736 | Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila],5M60_A Chain A, Beta-1,3-glucanase [Thermochaetoides thermophila] |
|
| 1.27e-118 | 165 | 911 | 26 | 744 | Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQN_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_A Chain A, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium],3EQO_B Chain B, Glucan 1,3-beta-glucosidase [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.26e-112 | 163 | 911 | 41 | 862 | Probable glucan endo-1,3-beta-glucosidase ARB_02077 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_02077 PE=1 SV=1 |
|
| 6.75e-92 | 164 | 914 | 47 | 776 | Glucan 1,3-beta-glucosidase OS=Cochliobolus carbonum OX=5017 GN=EXG1 PE=1 SV=1 |
|
| 8.70e-34 | 199 | 924 | 71 | 758 | Glucan endo-1,3-beta-glucosidase BGN13.1 OS=Trichoderma harzianum OX=5544 GN=bgn13.1 PE=1 SV=1 |
|
| 9.51e-07 | 185 | 360 | 4 | 178 | Alginate lyase 7 OS=Azotobacter vinelandii OX=354 GN=algE7 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
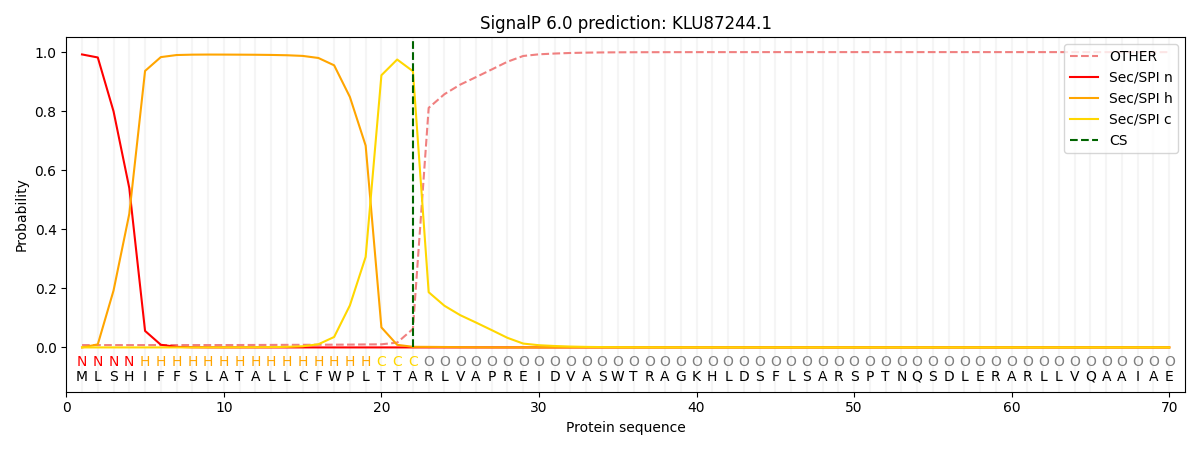
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.010135 | 0.989850 | CS pos: 22-23. Pr: 0.9353 |
