You are browsing environment: FUNGIDB
CAZyme Information: KLU85737.1
You are here: Home > Sequence: KLU85737.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Magnaporthiopsis poae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Magnaporthiopsis; Magnaporthiopsis poae | |||||||||||
| CAZyme ID | KLU85737.1 | |||||||||||
| CAZy Family | GH128 | |||||||||||
| CAZyme Description | acetyl-CoA carboxylase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH146 | 399 | 854 | 1.7e-143 | 0.8031809145129225 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 400342 | Glyco_hydro_127 | 1.78e-99 | 415 | 854 | 102 | 502 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| 226063 | COG3533 | 5.06e-64 | 434 | 948 | 130 | 587 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
| 404293 | Laminin_G_3 | 1.10e-29 | 184 | 343 | 1 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| 400342 | Glyco_hydro_127 | 5.82e-07 | 51 | 146 | 1 | 95 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| 214722 | LamGL | 0.001 | 201 | 340 | 1 | 133 | LamG-like jellyroll fold domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 31 | 982 | 55 | 1054 | |
| 0.0 | 8 | 982 | 12 | 1021 | |
| 0.0 | 8 | 982 | 12 | 1031 | |
| 0.0 | 8 | 981 | 12 | 1029 | |
| 0.0 | 8 | 981 | 12 | 1026 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.26e-65 | 423 | 854 | 152 | 559 | Chain AAA, Acetyl-CoA carboxylase, biotin carboxylase [Bacteroides thetaiotaomicron VPI-5482] |
|
| 1.96e-62 | 423 | 854 | 152 | 559 | Beta-L-arabinofuranosidase [Bacteroides thetaiotaomicron] |
|
| 1.61e-20 | 168 | 349 | 39 | 223 | Chain A, S-layer domain-containing protein [Acetivibrio thermocellus ATCC 27405] |
|
| 3.79e-17 | 415 | 853 | 128 | 543 | The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482],6EX6_B The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
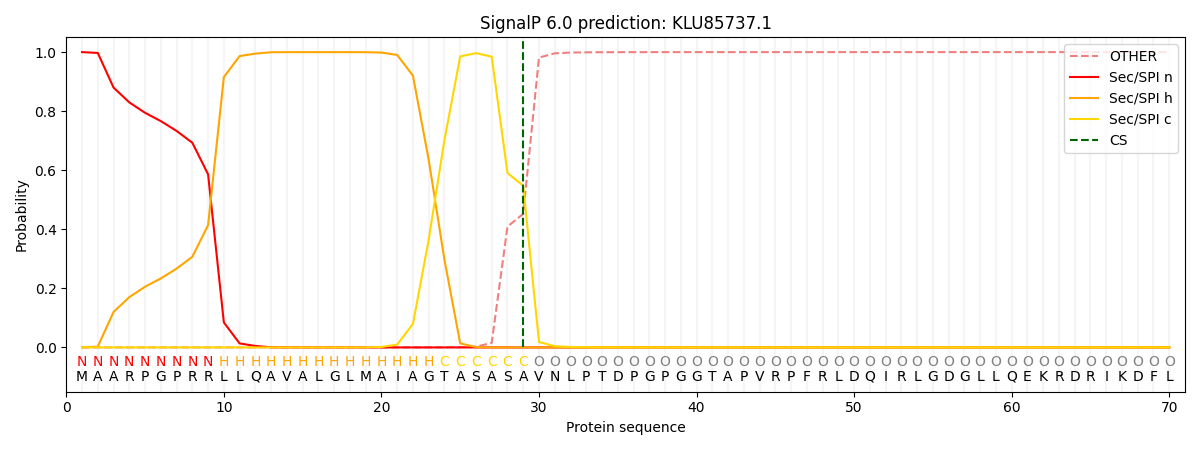
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000544 | 0.999422 | CS pos: 29-30. Pr: 0.5476 |
