You are browsing environment: FUNGIDB
CAZyme Information: KLU85393.1
You are here: Home > Sequence: KLU85393.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Magnaporthiopsis poae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Magnaporthaceae; Magnaporthiopsis; Magnaporthiopsis poae | |||||||||||
| CAZyme ID | KLU85393.1 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | 1,3-beta-glucan synthase component FKS1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2846259; End:2852287 Strand: + | |||||||||||
Full Sequence Download help
| MSGPQRPNRN DYDDGYGRNQ GGGGGGGGDY YQDDHSQQQQ YYDNGYDSGQ GGYGGQGGQG | 60 |
| GRNGDGYYDE SGYYNADPNN PYQQDGGYYD NNDQYQDDYY NNNGAYYDQG YDQGYNNPNG | 120 |
| RRGQGSEDDS ETFSDFTTGG PGGDYYGGGD PRYNSYNDGQ GGGRGYRPPS SQVSYGGNRS | 180 |
| SGASTPNYGM DYGGAAAAQR SREPYPAWTS DAQIPLSKEE VEDIFIDLCS KFGFQRDSMR | 240 |
| NMYDHLMTLL DSRASRMTPN QALLSLHADY IGGDNANYRK WYFAAHLDLD DAVGFANVKG | 300 |
| KGLKRKNKKK KGGDPQNEAD MLQDLEGDDS LEAAEYRWKT RMNRMSQHDR VRQLALYLLC | 360 |
| WGEANQVRFM PECLCFIFKC ADDYLNSPAC QNLVEPVEEF TFLNQVITPL YQYCRDQGYE | 420 |
| IIDGVYVRRE RDHNKIIGYD DCNQLFWYPE GIERIVLEDK TKLTDVPPAE RYLKLKDVNW | 480 |
| KKCFFKTYKE TRSWFHLITN FNRIWIIHLT MWWYYTAFNA PTLLVPNYEQ QANNVPPNSV | 540 |
| MWSICGLGGG IACLIQIMAT ICEWMYVPRR WAGAQHLTKR LLFLLALFVI NVGPAVYIFM | 600 |
| PAKDLATFEA RQKSQVALII GIVHFFISVI TFIFFTIMPL GGLFGSYLTK NSRRYVASQT | 660 |
| FTASYPRLAG NDMVLSYLLW LIVFGAKLGE SYGYLALSFR DPVRYLQIMD IDCRGDKLFP | 720 |
| PAMQNMLCKL QPKFLLCLMA FTDLIFFFLD TYLWYVLINT AVSIARSFYL GASILTPWRN | 780 |
| IFSRLPKRIY SKVLATTDME IKYKPKVLIS QIWNAIVISM YREHLLAIDH VQKLLYHQVP | 840 |
| SEQEGKRTLR APTFFVSQED HSFKTEFFPA QSEAERRISF FAQSLSIPIP EPLPVDNMPT | 900 |
| FTVLIPHYSE KILLSLREII REDEPYSRVT LLEYLKQLYP HEWDCFVKDT KILADETSQF | 960 |
| NGDGEKDEKD TAKSKIDDLP FYCIGFKSSA PEYTLRTRIW SSLRSQTLYR TISGFMNYSR | 1020 |
| AIKLLYRVEN PEVVQMFGGN SEKLERELER MARRKFKLVV SMQRFAKFKK EEMENAEFLL | 1080 |
| RAYPDLQIAY LDEEAPLNEG DEPRIYSALI DGHSEIMENG VRRPKFRIQL SGNPILGDGK | 1140 |
| SDNQNHSIIF YRGEYIQLID ANQDNYLEEC LKIRSVLAEF EEMKVDNTSP YTPGVKNPVK | 1200 |
| SPVAILGARE YIFSENIGIL GDVAAGKEQT FGTLFARTLA QIGGKLHYGH PDFLNGIFMT | 1260 |
| TRGGVSKAQK GLHLNEDIYA GMNAVLRGGR IKHCEYYQCG KGRDLGFGSI LNFTTKIGTG | 1320 |
| MGEQMLSREY YYLGTQLPLD RFLSFYYAHP GFHINNIFIM LSIQMFMITL LNIGALKHET | 1380 |
| IPCNYDRNVP ITDELFPTGC QNTEALTDWV FRSVLSIIFV LCLSYVPLIV QELFERGVSR | 1440 |
| AAFRLAKQIC SLSPLFEVFV CQIYANAVHN NLSFGGARYI GTGRGFATAR IPFGVLYSRF | 1500 |
| AAPSIYFGAR LLMMLLFATV TIFQGALVYF WITLLALVIS PFLYNPHQFA WNDFFIDYRD | 1560 |
| YLRWLSRGNS RSHASSWIAY CRLSRTRLTG YKRKAAGDPS AKMSADVPRA SFTNVFFSEI | 1620 |
| VSPFLLVIVT VVPFMYLNAQ TGVLPDRNPN QTIAPTGALL RIAVIALAPI GVNAVVLLIM | 1680 |
| FFMACFMGPL LSMCCKKFGS VLAAIAHALA VIMLLIFCEV LFVLERFSFA RTLSGMIAVV | 1740 |
| AIQRFLLKLI VCLGLTREFK TDQANIAFWT GKWYSMGWHT ISQPAREFLC KITELSFFAS | 1800 |
| DFILGHVLLF FMLPVIIIPQ VDKLHSTMLF WLRPSRQIRP PIYSMKQSKL RQRRVIRYAI | 1860 |
| LYFGLLVVFV ALIGGPVFLG GQPTLVDTIT DPIGKISPIK SFGLFQSKVA SNNNTNKTEA | 1920 |
| TGTRMPGYSG VRATTSSAAT TGLGARSVVL I | 1951 |
Enzyme Prediction help
| EC | 2.4.1.34:56 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT48 | 867 | 1644 | 0 | 0.9878213802435724 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396784 | Glucan_synthase | 0.0 | 867 | 1691 | 1 | 819 | 1,3-beta-glucan synthase component. This family consists of various 1,3-beta-glucan synthase components including Gls1, Gls2 and Gls3 from yeast. 1,3-beta-glucan synthase EC:2.4.1.34 also known as callose synthase catalyzes the formation of a beta-1,3-glucan polymer that is a major component of the fungal cell wall. The reaction catalyzed is:- UDP-glucose + {(1,3)-beta-D-glucosyl}(N) <=> UDP + {(1,3)-beta-D-glucosyl}(N+1). |
| 405046 | FKS1_dom1 | 4.38e-50 | 349 | 458 | 1 | 111 | 1,3-beta-glucan synthase subunit FKS1, domain-1. The FKS1_dom1 domain is likely to be the 'Class I' region just N-terminal to the first set of transmembrane helices that is involved in 1,3-beta-glucan synthesis itself. This family is found on proteins with family Glucan_synthase, pfam02364. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QBZ62345.1|GT48 | 0.0 | 5 | 1927 | 8 | 1910 |
| AEO59893.1|GT48 | 0.0 | 75 | 1950 | 64 | 1933 |
| AEO68099.1|GT48 | 0.0 | 108 | 1923 | 97 | 1907 |
| UNI17292.1|GT48 | 0.0 | 75 | 1942 | 62 | 1929 |
| UQC87088.1|GH17|GT48 | 0.0 | 76 | 1937 | 76 | 1930 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P40989|FKS2_YEAST | 0.0 | 83 | 1876 | 48 | 1837 | 1,3-beta-glucan synthase component GSC2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=GSC2 PE=1 SV=2 |
| sp|A2QLK4|FKS1_ASPNC | 0.0 | 75 | 1876 | 50 | 1844 | 1,3-beta-glucan synthase component FKS1 OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=fksA PE=3 SV=1 |
| sp|Q04952|FKS3_YEAST | 0.0 | 205 | 1915 | 15 | 1763 | 1,3-beta-glucan synthase component FKS3 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS3 PE=1 SV=1 |
| sp|P38631|FKS1_YEAST | 0.0 | 111 | 1876 | 66 | 1818 | 1,3-beta-glucan synthase component FKS1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=FKS1 PE=1 SV=2 |
| sp|O93927|FKS1_CRYNH | 0.0 | 168 | 1877 | 104 | 1780 | 1,3-beta-glucan synthase component FKS1 OS=Cryptococcus neoformans var. grubii serotype A (strain H99 / ATCC 208821 / CBS 10515 / FGSC 9487) OX=235443 GN=FKS1 PE=3 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000065 | 0.000000 |
TMHMM Annotations download full data without filtering help
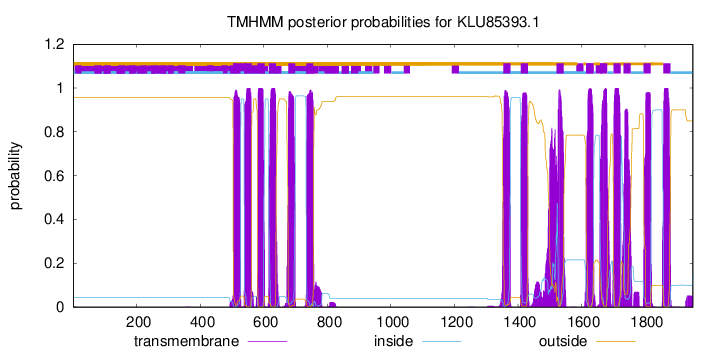
| Start | End |
|---|---|
| 504 | 526 |
| 539 | 561 |
| 581 | 603 |
| 616 | 638 |
| 677 | 699 |
| 734 | 756 |
| 1354 | 1376 |
| 1409 | 1431 |
| 1522 | 1544 |
| 1615 | 1637 |
| 1659 | 1681 |
| 1701 | 1723 |
| 1733 | 1755 |
| 1796 | 1818 |
| 1858 | 1880 |
