You are browsing environment: FUNGIDB
CAZyme Information: KIW19871.1
You are here: Home > Sequence: KIW19871.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Exophiala spinifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala spinifera | |||||||||||
| CAZyme ID | KIW19871.1 | |||||||||||
| CAZy Family | GT2|GT2 | |||||||||||
| CAZyme Description | Glyco_trans_2-like domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A0D1YX53] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT2 | 512 | 724 | 9.9e-49 | 0.9796954314720813 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 404513 | Glyco_trans_2_3 | 6.55e-46 | 512 | 727 | 1 | 194 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| 224136 | BcsA | 4.40e-16 | 502 | 776 | 131 | 386 | Glycosyltransferase, catalytic subunit of cellulose synthase and poly-beta-1,6-N-acetylglucosamine synthase [Cell motility]. |
| 133045 | CESA_like | 3.74e-11 | 510 | 620 | 79 | 180 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
| 225494 | MdoH | 2.06e-09 | 497 | 824 | 232 | 556 | Membrane glycosyltransferase [Cell wall/membrane/envelope biogenesis, Carbohydrate transport and metabolism]. |
| 224137 | GT2 | 2.67e-06 | 476 | 636 | 59 | 211 | Glycosyltransferase, GT2 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 64 | 899 | 69 | 887 | |
| 0.0 | 46 | 900 | 48 | 883 | |
| 6.47e-318 | 46 | 900 | 48 | 883 | |
| 2.19e-316 | 66 | 900 | 72 | 888 | |
| 1.74e-313 | 46 | 900 | 48 | 900 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.45e-06 | 513 | 704 | 241 | 435 | Glucans biosynthesis glucosyltransferase H OS=Xanthomonas euvesicatoria OX=456327 GN=opgH PE=3 SV=1 |
|
| 2.46e-06 | 513 | 704 | 241 | 435 | Glucans biosynthesis glucosyltransferase H OS=Xanthomonas campestris pv. vesicatoria (strain 85-10) OX=316273 GN=opgH PE=3 SV=1 |
|
| 3.23e-06 | 513 | 704 | 241 | 435 | Glucans biosynthesis glucosyltransferase H OS=Xanthomonas oryzae pv. oryzae (strain MAFF 311018) OX=342109 GN=opgH PE=3 SV=1 |
|
| 3.23e-06 | 513 | 704 | 241 | 435 | Glucans biosynthesis glucosyltransferase H OS=Xanthomonas axonopodis pv. citri (strain 306) OX=190486 GN=opgH PE=3 SV=2 |
|
| 4.25e-06 | 510 | 695 | 236 | 425 | Glucans biosynthesis glucosyltransferase H OS=Xanthomonas campestris pv. campestris (strain B100) OX=509169 GN=opgH PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000072 | 0.000001 |
TMHMM Annotations download full data without filtering help
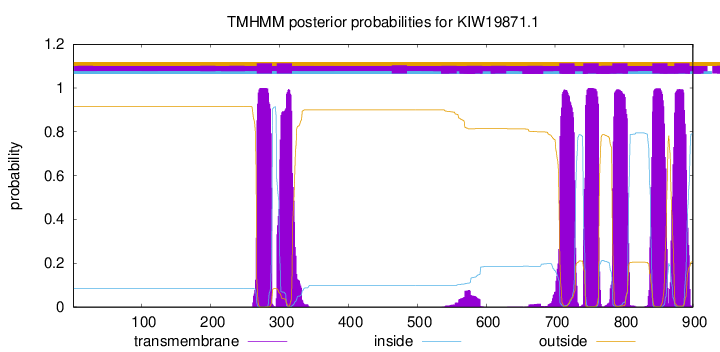
| Start | End |
|---|---|
| 267 | 289 |
| 296 | 318 |
| 706 | 728 |
| 741 | 763 |
| 783 | 805 |
| 841 | 863 |
| 873 | 895 |
