You are browsing environment: FUNGIDB
CAZyme Information: KIW19001.1
You are here: Home > Sequence: KIW19001.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Exophiala spinifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala spinifera | |||||||||||
| CAZyme ID | KIW19001.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Alpha-1,3-glucan synthase [Source:UniProtKB/TrEMBL;Acc:A0A0D1YUT5] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.183:36 | 2.4.1.-:11 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340822 | GT5_Glycogen_synthase_DULL1-like | 2.04e-89 | 9 | 405 | 60 | 459 | Glycogen synthase GlgA and similar proteins. This family is most closely related to the GT5 family of glycosyltransferases. Glycogen synthase (EC:2.4.1.21) catalyzes the formation and elongation of the alpha-1,4-glucose backbone using ADP-glucose, the second and key step of glycogen biosynthesis. This family includes starch synthases of plants, such as DULL1 in Zea mays and glycogen synthases of various organisms. |
| 223374 | GlgA | 1.06e-22 | 11 | 356 | 65 | 403 | Glycogen synthase [Carbohydrate transport and metabolism]. |
| 223374 | GlgA | 6.33e-20 | 926 | 1345 | 113 | 487 | Glycogen synthase [Carbohydrate transport and metabolism]. |
| 237610 | PRK14099 | 5.62e-17 | 31 | 356 | 84 | 404 | glycogen synthase GlgA. |
| 234809 | glgA | 4.22e-16 | 17 | 356 | 63 | 391 | glycogen synthase GlgA. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1367 | 1226 | 2467 | |
| 0.0 | 1 | 1367 | 1226 | 2467 | |
| 0.0 | 1 | 1367 | 1224 | 2460 | |
| 0.0 | 1 | 1367 | 1212 | 2422 | |
| 0.0 | 1 | 1367 | 1212 | 2422 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.43e-12 | 77 | 356 | 130 | 401 | Chain A, Glycogen synthase [Escherichia coli] |
|
| 1.47e-12 | 77 | 356 | 130 | 401 | Crystal Structure of Wild-type E.coli GS in complex with ADP and Glucose(wtGSb) [Escherichia coli],2R4T_A Crystal Structure of Wild-type E.coli GS in Complex with ADP and Glucose(wtGSc) [Escherichia coli],2R4U_A Crystal Structure of Wild-type E.coli GS in complex with ADP and Glucose(wtGSd) [Escherichia coli],3GUH_A Crystal Structure of Wild-type E.coli GS in complex with ADP and DGM [Escherichia coli K-12] |
|
| 1.74e-12 | 83 | 356 | 170 | 458 | Granule Bound Starch Synthase from Cyanobacterium sp. CLg1 bound to acarbose and ADP [Cyanobacterium sp. CLg1],6GNF_B Granule Bound Starch Synthase from Cyanobacterium sp. CLg1 bound to acarbose and ADP [Cyanobacterium sp. CLg1],6GNF_C Granule Bound Starch Synthase from Cyanobacterium sp. CLg1 bound to acarbose and ADP [Cyanobacterium sp. CLg1] |
|
| 3.09e-11 | 75 | 356 | 179 | 478 | Granule Bound Starch Synthase I from Cyanophora paradoxa bound to acarbose and ADP [Cyanophora paradoxa],6GNG_B Granule Bound Starch Synthase I from Cyanophora paradoxa bound to acarbose and ADP [Cyanophora paradoxa] |
|
| 1.08e-10 | 75 | 356 | 151 | 436 | Crystal Structure of Rice Granule bound Starch Synthase I Catalytic Domain [Oryza sativa Japonica Group],3VUF_A Crystal Structure of Rice Granule bound Starch Synthase I Catalytic Domain in Complex with ADP [Oryza sativa Japonica Group] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.77e-302 | 5 | 1367 | 1201 | 2410 | Cell wall alpha-1,3-glucan synthase ags1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=ags1 PE=1 SV=3 |
|
| 5.13e-286 | 7 | 1367 | 1198 | 2358 | Cell wall alpha-1,3-glucan synthase mok13 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok13 PE=3 SV=2 |
|
| 1.90e-271 | 7 | 1367 | 1205 | 2397 | Cell wall alpha-1,3-glucan synthase mok11 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok11 PE=3 SV=2 |
|
| 9.20e-230 | 5 | 1367 | 312 | 1369 | Cell wall alpha-1,3-glucan synthase mok14 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok14 PE=1 SV=1 |
|
| 1.04e-218 | 8 | 1364 | 1232 | 2350 | Cell wall alpha-1,3-glucan synthase mok12 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=mok12 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000074 | 0.000000 |
TMHMM Annotations download full data without filtering help
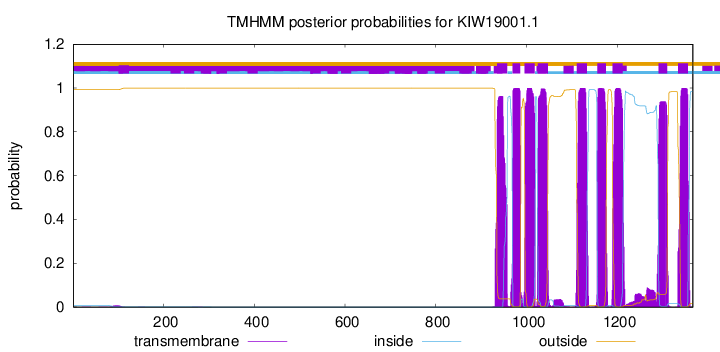
| Start | End |
|---|---|
| 935 | 957 |
| 969 | 986 |
| 996 | 1018 |
| 1025 | 1047 |
| 1111 | 1133 |
| 1156 | 1175 |
| 1190 | 1212 |
| 1291 | 1310 |
| 1334 | 1356 |
