You are browsing environment: FUNGIDB
CAZyme Information: KIW13484.1
You are here: Home > Sequence: KIW13484.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
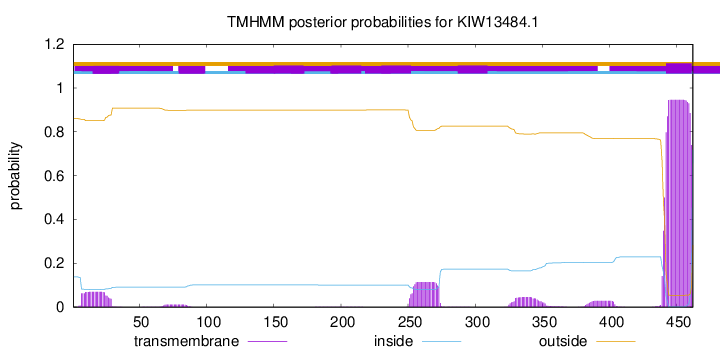
TMHMM annotations
Basic Information help
| Species | Exophiala spinifera | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala spinifera | |||||||||||
| CAZyme ID | KIW13484.1 | |||||||||||
| CAZy Family | GH13 | |||||||||||
| CAZyme Description | Mannan endo-1,6-alpha-mannosidase [Source:UniProtKB/TrEMBL;Acc:A0A0D2BQR9] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.101:1 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 27 | 400 | 4e-103 | 0.9413407821229051 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397638 | Glyco_hydro_76 | 1.22e-167 | 31 | 390 | 1 | 348 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.89e-164 | 1 | 461 | 3 | 450 | |
| 3.79e-159 | 18 | 461 | 21 | 451 | |
| 5.00e-157 | 6 | 461 | 8 | 450 | |
| 2.02e-156 | 6 | 461 | 8 | 450 | |
| 1.07e-151 | 6 | 446 | 4 | 435 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.94e-106 | 25 | 420 | 29 | 425 | Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY1_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY2_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY5_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY6_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495],6RY7_A Chain A, Mannan endo-1,6-alpha-mannosidase [Thermochaetoides thermophila DSM 1495] |
|
| 7.32e-10 | 70 | 336 | 88 | 325 | Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
|
| 1.26e-08 | 70 | 336 | 89 | 326 | Chain A, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
|
| 4.98e-08 | 150 | 274 | 129 | 252 | Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 6.94e-08 | 70 | 336 | 89 | 326 | An inactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.85e-122 | 13 | 461 | 7 | 448 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DCW1 PE=1 SV=1 |
|
| 3.46e-119 | 7 | 461 | 5 | 445 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=DCW1 PE=3 SV=1 |
|
| 1.19e-118 | 17 | 462 | 14 | 452 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=DCW1 PE=1 SV=1 |
|
| 1.18e-115 | 22 | 462 | 22 | 451 | Mannan endo-1,6-alpha-mannosidase DCW1 OS=Ashbya gossypii (strain ATCC 10895 / CBS 109.51 / FGSC 9923 / NRRL Y-1056) OX=284811 GN=DCW1 PE=3 SV=2 |
|
| 6.83e-105 | 20 | 459 | 25 | 455 | Mannan endo-1,6-alpha-mannosidase DFG5 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=DFG5 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000275 | 0.999682 | CS pos: 21-22. Pr: 0.9769 |