You are browsing environment: FUNGIDB
CAZyme Information: KIW01591.1
You are here: Home > Sequence: KIW01591.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Verruconis gallopava | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Dothideomycetes; ; Sympoventuriaceae; Verruconis; Verruconis gallopava | |||||||||||
| CAZyme ID | KIW01591.1 | |||||||||||
| CAZy Family | GH115 | |||||||||||
| CAZyme Description | GMC_OxRdtase_N domain-containing protein [Source:UniProtKB/TrEMBL;Acc:A0A0D1XH98] | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 433 | 1050 | 5e-95 | 0.9947183098591549 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 1.54e-57 | 427 | 1043 | 1 | 528 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 1.39e-56 | 431 | 1043 | 3 | 526 | choline dehydrogenase; Validated |
| 398739 | GMC_oxred_C | 1.18e-27 | 909 | 1043 | 5 | 142 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 215420 | PLN02785 | 1.99e-18 | 697 | 1031 | 234 | 559 | Protein HOTHEAD |
| 399033 | CFEM | 7.12e-12 | 26 | 90 | 1 | 66 | CFEM domain. This fungal specific cysteine rich domain is found in some proteins with proposed roles in fungal pathogenesis. The structure of the CFEM domain containing protein 'Surface antigen protein 2' from Candida albicans has been solved. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.90e-301 | 424 | 1052 | 25 | 670 | |
| 1.62e-298 | 432 | 1054 | 47 | 676 | |
| 8.56e-258 | 432 | 1056 | 48 | 683 | |
| 3.49e-253 | 432 | 1055 | 48 | 678 | |
| 3.23e-245 | 433 | 1055 | 38 | 668 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.55e-33 | 433 | 1048 | 40 | 597 | Crystal structure of pyranose dehydrogenase from Agaricus meleagris, wildtype [Leucoagaricus meleagris] |
|
| 5.96e-29 | 433 | 1058 | 7 | 570 | Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis] |
|
| 1.06e-28 | 430 | 1058 | 4 | 570 | Chain A, Fatty acid photodecarboxylase, chloroplastic [Chlorella variabilis],6ZH7_B Chain B, Fatty acid photodecarboxylase, chloroplastic [Chlorella variabilis] |
|
| 1.88e-28 | 433 | 1058 | 7 | 570 | Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis] |
|
| 2.50e-28 | 433 | 1058 | 7 | 570 | Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis],6YRV_AAA Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis],6YRX_AAA Chain AAA, Fatty acid Photodecarboxylase [Chlorella variabilis],6YRZ_AAA Chain AAA, Fatty acid photodecarboxylase, chloroplastic [Chlorella variabilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.09e-41 | 432 | 1048 | 41 | 573 | Choline dehydrogenase, mitochondrial OS=Mus musculus OX=10090 GN=Chdh PE=1 SV=1 |
|
| 4.60e-41 | 434 | 1049 | 5 | 531 | Oxygen-dependent choline dehydrogenase OS=Photobacterium profundum (strain SS9) OX=298386 GN=betA PE=3 SV=1 |
|
| 6.18e-41 | 433 | 1046 | 6 | 533 | Oxygen-dependent choline dehydrogenase OS=Pseudomonas syringae pv. tomato (strain ATCC BAA-871 / DC3000) OX=223283 GN=betA PE=3 SV=1 |
|
| 1.34e-40 | 434 | 1049 | 5 | 531 | Oxygen-dependent choline dehydrogenase OS=Vibrio vulnificus (strain YJ016) OX=196600 GN=betA PE=3 SV=1 |
|
| 1.48e-40 | 433 | 1046 | 2 | 529 | Oxygen-dependent choline dehydrogenase OS=Yersinia pseudotuberculosis serotype IB (strain PB1/+) OX=502801 GN=betA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
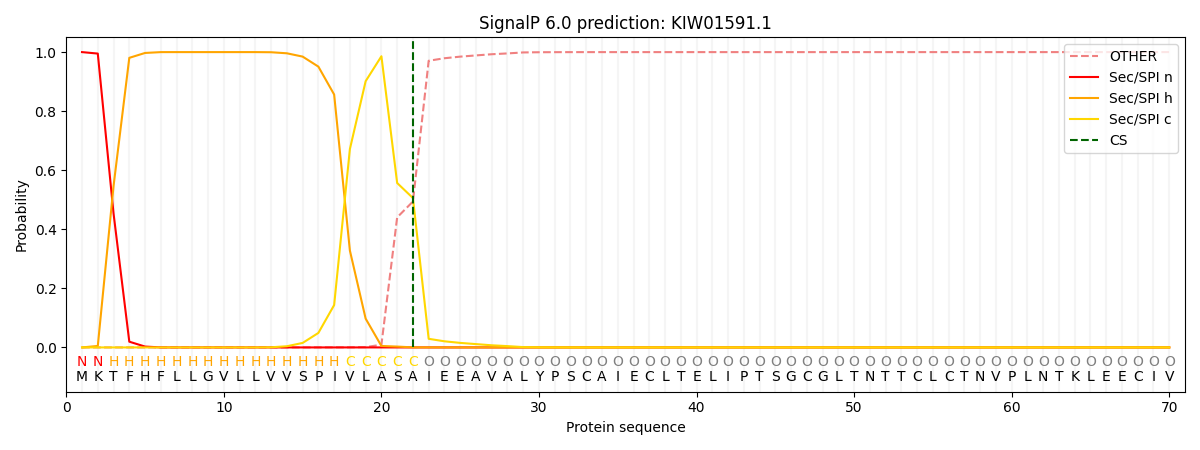
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000187 | 0.999795 | CS pos: 22-23. Pr: 0.5058 |
TMHMM Annotations download full data without filtering help
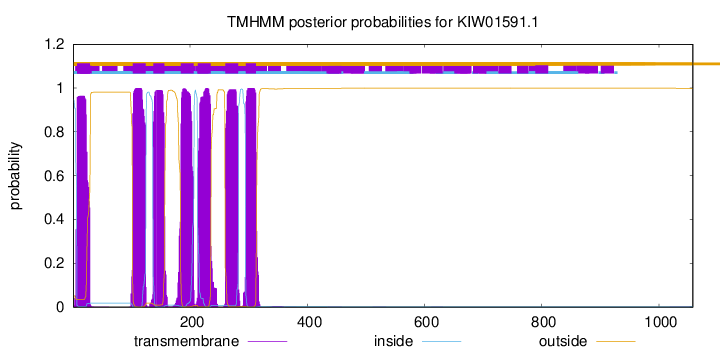
| Start | End |
|---|---|
| 7 | 29 |
| 102 | 124 |
| 137 | 156 |
| 184 | 206 |
| 213 | 235 |
| 260 | 282 |
| 295 | 312 |
