You are browsing environment: FUNGIDB
CAZyme Information: KIS70137.1
You are here: Home > Sequence: KIS70137.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Ustilago maydis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Ustilaginomycetes; ; Ustilaginaceae; Ustilago; Ustilago maydis | |||||||||||
| CAZyme ID | KIS70137.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.131:1 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340835 | GT4_ALG11-like | 0.0 | 343 | 772 | 2 | 416 | alpha-1,2-mannosyltransferase ALG11 and similar proteins. This family is most closely related to the GT4 family of glycosyltransferases. ALG11 in yeast is involved in adding the final 1,2-linked Man to the Man5GlcNAc2-PP-Dol synthesized on the cytosolic face of the ER. The deletion analysis of ALG11 was shown to block the early steps of core biosynthesis that takes place on the cytoplasmic face of the ER and lead to a defect in the assembly of lipid-linked oligosaccharides. |
| 215511 | PLN02949 | 1.23e-126 | 344 | 772 | 36 | 446 | transferase, transferring glycosyl groups |
| 406369 | ALG11_N | 9.42e-118 | 342 | 549 | 1 | 207 | ALG11 mannosyltransferase N-terminus. |
| 398767 | EBP | 1.02e-24 | 76 | 187 | 1 | 113 | Emopamil binding protein. Emopamil binding protein (EBP) is as a gene that encodes a non-glycosylated type I integral membrane protein of endoplasmic reticulum and shows high level expression in epithelial tissues. The EBP protein has emopamil binding domains, including the sterol acceptor site and the catalytic centre, which show Delta8-Delta7 sterol isomerase activity. Human sterol isomerase, a homolog of mouse EBP, is suggested not only to play a role in cholesterol biosynthesis, but also to affect lipoprotein internalisation. In humans, mutations of EBP are known to cause the genetic disorder of X-linked dominant chondrodysplasia punctata (CDPX2). This syndrome of humans is lethal in most males, and affected females display asymmetric hyperkeratotic skin and skeletal abnormalities. |
| 340834 | GT4_ALG2-like | 1.05e-16 | 344 | 772 | 3 | 388 | alpha-1,3/1,6-mannosyltransferase ALG2 and similar proteins. This family is most closely related to the GT4 family of glycosyltransferases. ALG2, a 1,3-mannosyltransferase, in yeast catalyzes the mannosylation of Man(2)GlcNAc(2)-dolichol diphosphate and Man(1)GlcNAc(2)-dolichol diphosphate to form Man(3)GlcNAc(2)-dolichol diphosphate. A deficiency of this enzyme causes an abnormal accumulation of Man1GlcNAc2-PP-dolichol and Man2GlcNAc2-PP-dolichol, which is associated with a type of congenital disorders of glycosylation (CDG), designated CDG-Ii, in humans. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 789 | 1 | 782 | |
| 0.0 | 1 | 789 | 1 | 786 | |
| 0.0 | 1 | 789 | 1 | 842 | |
| 5.47e-267 | 145 | 789 | 26 | 661 | |
| 1.64e-163 | 1 | 382 | 1 | 392 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.54e-12 | 40 | 196 | 71 | 224 | Structure of EBP and tamoxifen [Homo sapiens],6OHU_B Structure of EBP and tamoxifen [Homo sapiens],6OHU_C Structure of EBP and tamoxifen [Homo sapiens] |
|
| 2.74e-10 | 40 | 196 | 71 | 224 | Structure of EBP and U18666A [Homo sapiens],6OHT_B Structure of EBP and U18666A [Homo sapiens],6OHT_C Structure of EBP and U18666A [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.56e-94 | 324 | 761 | 45 | 463 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Xenopus laevis OX=8355 GN=alg11 PE=2 SV=2 |
|
| 1.06e-92 | 322 | 772 | 45 | 474 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Xenopus tropicalis OX=8364 GN=alg11 PE=2 SV=1 |
|
| 1.31e-92 | 326 | 773 | 43 | 453 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=alg11 PE=3 SV=1 |
|
| 7.89e-88 | 340 | 760 | 61 | 468 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Homo sapiens OX=9606 GN=ALG11 PE=1 SV=2 |
|
| 3.01e-87 | 340 | 760 | 61 | 468 | GDP-Man:Man(3)GlcNAc(2)-PP-Dol alpha-1,2-mannosyltransferase OS=Pongo abelii OX=9601 GN=ALG11 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
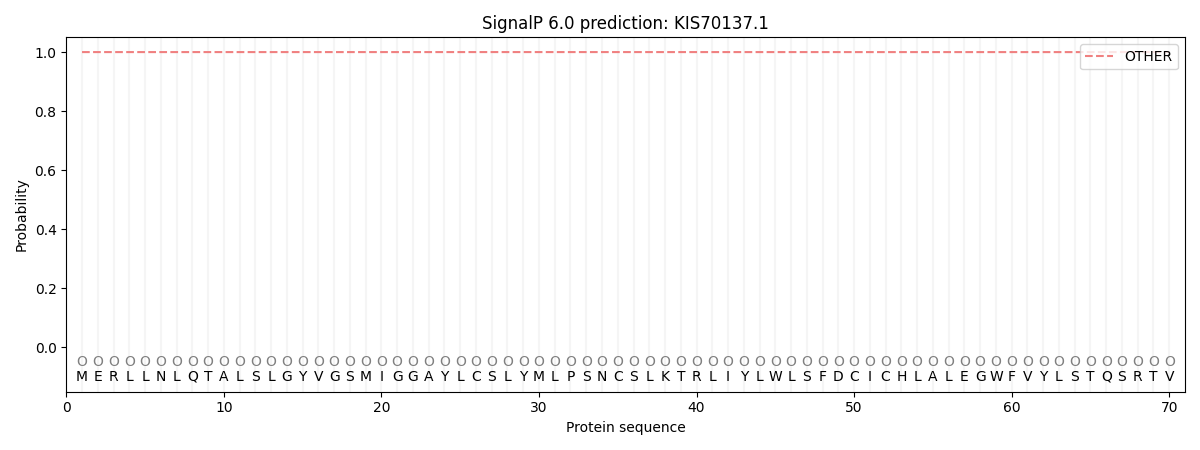
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000054 | 0.000000 |
TMHMM Annotations download full data without filtering help
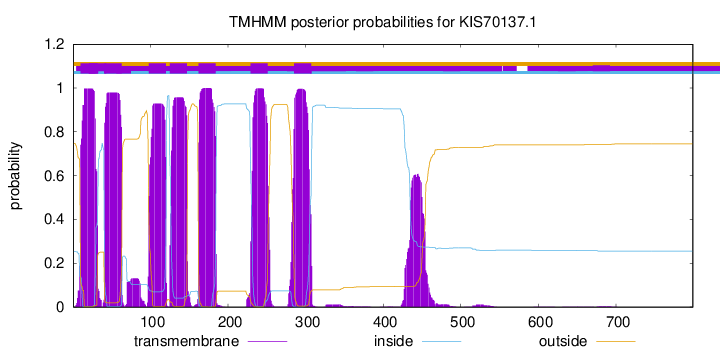
| Start | End |
|---|---|
| 10 | 32 |
| 41 | 63 |
| 98 | 120 |
| 125 | 147 |
| 162 | 184 |
| 229 | 251 |
| 285 | 307 |
