You are browsing environment: FUNGIDB
CAZyme Information: KIS69676.1
You are here: Home > Sequence: KIS69676.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
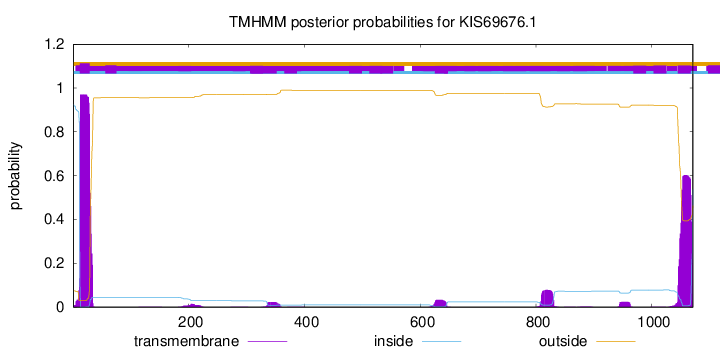
TMHMM annotations
Basic Information help
| Species | Ustilago maydis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Ustilaginomycetes; ; Ustilaginaceae; Ustilago; Ustilago maydis | |||||||||||
| CAZyme ID | KIS69676.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH105 | 779 | 991 | 5.4e-25 | 0.6265060240963856 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 400034 | Glyco_hydro_88 | 6.46e-14 | 780 | 957 | 93 | 263 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| 226678 | YesR | 4.12e-11 | 812 | 1008 | 132 | 329 | Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism]. |
| 341250 | LC-FACS_euk | 1.02e-10 | 397 | 586 | 228 | 400 | Eukaryotic long-chain fatty acid CoA synthetase (LC-FACS). The members of this family are eukaryotic fatty acid CoA synthetases that activate fatty acids with chain lengths of 12 to 20. LC-FACS catalyzes the formation of fatty acyl-CoA in a two-step reaction: the formation of a fatty acyl-AMP molecule as an intermediate, and the formation of a fatty acyl-CoA. This is a required step before free fatty acids can participate in most catabolic and anabolic reactions. Organisms tend to have multiple isoforms of LC-FACS genes with multiple splice variants. For example, nine genes are found in Arabidopsis and six genes are expressed in mammalian cells. |
| 341294 | LC_FACS_euk1 | 1.16e-08 | 410 | 586 | 223 | 375 | Eukaryotic long-chain fatty acid CoA synthetase (LC-FACS), including fungal proteins. The members of this family are eukaryotic fatty acid CoA synthetases (EC 6.2.1.3) that activate fatty acids with chain lengths of 12 to 20 and includes fungal proteins. They act on a wide range of long-chain saturated and unsaturated fatty acids, but the enzymes from different tissues show some variation in specificity. LC-FACS catalyzes the formation of fatty acyl-CoA in a two-step reaction: the formation of a fatty acyl-AMP molecule as an intermediate, and the formation of a fatty acyl-CoA. This is a required step before free fatty acids can participate in most catabolic and anabolic reactions. Organisms tend to have multiple isoforms of LC-FACS genes with multiple splice variants. For example, nine genes are found in Arabidopsis and six genes are expressed in mammalian cells. In Schizosaccharomyces pombe, lcf1 gene encodes a new fatty acyl-CoA synthetase that preferentially recognizes myristic acid as a substrate. |
| 223953 | FAA1 | 1.31e-06 | 408 | 589 | 322 | 468 | Long-chain acyl-CoA synthetase (AMP-forming) [Lipid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 1070 | 1 | 1064 | |
| 0.0 | 1 | 1070 | 1 | 1064 | |
| 0.0 | 2 | 1064 | 1 | 1062 | |
| 0.0 | 1 | 1071 | 1 | 1068 | |
| 0.0 | 1 | 1071 | 1 | 1067 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
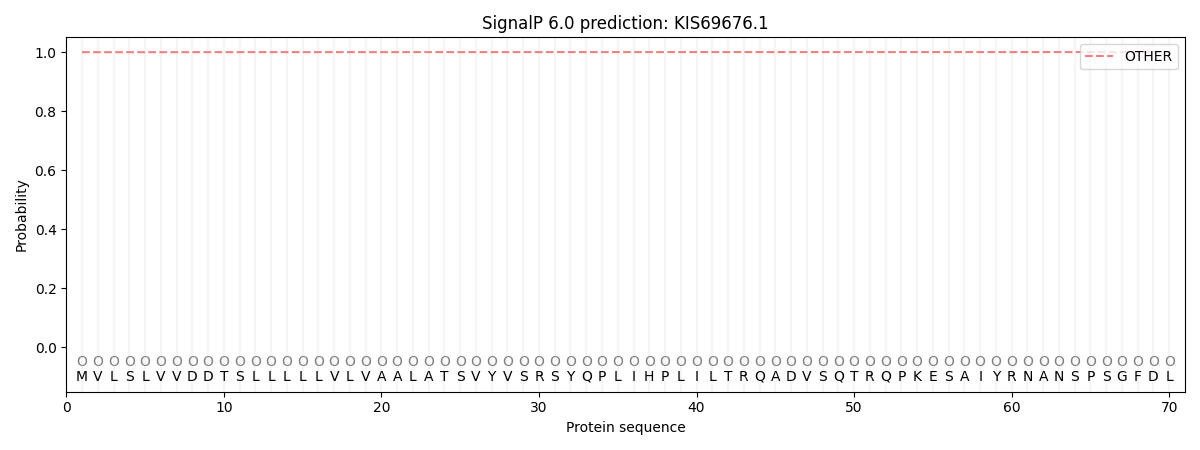
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999683 | 0.000365 |