You are browsing environment: FUNGIDB
CAZyme Information: KID66592.1
You are here: Home > Sequence: KID66592.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
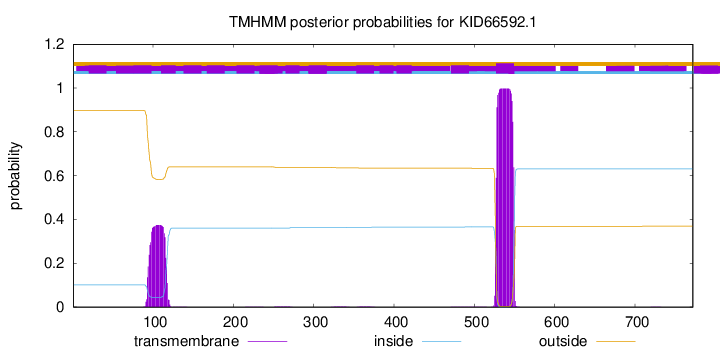
TMHMM annotations
Basic Information help
| Species | Metarhizium anisopliae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Clavicipitaceae; Metarhizium; Metarhizium anisopliae | |||||||||||
| CAZyme ID | KID66592.1 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | cystathionine beta-lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE4 | 571 | 702 | 4.4e-23 | 0.9615384615384616 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 99738 | CGS_like | 4.97e-94 | 34 | 406 | 2 | 369 | CGS_like: Cystathionine gamma-synthase is a PLP dependent enzyme and catalyzes the committed step of methionine biosynthesis. This pathway is unique to microorganisms and plants, rendering the enzyme an attractive target for the development of antimicrobials and herbicides. This subgroup also includes cystathionine gamma-lyases (CGL), O-acetylhomoserine sulfhydrylases and O-acetylhomoserine thiol lyases. CGL's are very similar to CGS's. Members of this group are widely distributed among all three forms of life. |
| 223699 | MetC | 9.94e-91 | 11 | 409 | 1 | 396 | Cystathionine beta-lyase/cystathionine gamma-synthase [Amino acid transport and metabolism]. |
| 200581 | CE4_NodB_like_2 | 4.71e-81 | 576 | 762 | 1 | 190 | Catalytic NodB homology domain of uncharacterized chitin deacetylases and hypothetical proteins. This family includes some uncharacterized chitin deacetylases and hypothetical proteins, mainly from eukaryotes. Although their biological function is unknown, members in this family show high sequence homology to the catalytic NodB homology domain of Colletotrichum lindemuthianum chitin deacetylase (endo-chitin de-N-acetylase, ClCDA, EC 3.5.1.41), which catalyzes the hydrolysis of N-acetamido groups of N-acetyl-D-glucosamine residues in chitin, converting it to chitosan in fungal cell walls. Like ClCDA, this family is a member the carbohydrate esterase 4 (CE4) superfamily. |
| 395837 | Cys_Met_Meta_PP | 4.51e-68 | 19 | 406 | 1 | 376 | Cys/Met metabolism PLP-dependent enzyme. This family includes enzymes involved in cysteine and methionine metabolism. The following are members: Cystathionine gamma-lyase, Cystathionine gamma-synthase, Cystathionine beta-lyase, Methionine gamma-lyase, OAH/OAS sulfhydrylase, O-succinylhomoserine sulfhydrylase All of these members participate is slightly different reactions. All these enzymes use PLP (pyridoxal-5'-phosphate) as a cofactor. |
| 236104 | PRK07811 | 4.25e-54 | 10 | 406 | 5 | 387 | cystathionine gamma-synthase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 6 | 772 | 5 | 637 | |
| 0.0 | 6 | 772 | 4 | 690 | |
| 1.70e-281 | 12 | 772 | 11 | 684 | |
| 2.21e-129 | 505 | 772 | 1 | 271 | |
| 2.69e-124 | 505 | 772 | 1 | 269 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.40e-42 | 19 | 410 | 9 | 393 | Crystal Structure of cystathionine gamma-lyase from yeast [Saccharomyces cerevisiae],1N8P_B Crystal Structure of cystathionine gamma-lyase from yeast [Saccharomyces cerevisiae],1N8P_C Crystal Structure of cystathionine gamma-lyase from yeast [Saccharomyces cerevisiae],1N8P_D Crystal Structure of cystathionine gamma-lyase from yeast [Saccharomyces cerevisiae] |
|
| 9.32e-40 | 80 | 404 | 58 | 392 | crystal structure of cystathionine gamma lyase (XometC) from Xanthomonas oryzae pv. oryzae in complex with E-site serine, A-site external aldimine structure with serine and A-site external aldimine structure with aminoacrylate intermediates [Xanthomonas oryzae pv. oryzae KACC 10331],4IY7_B crystal structure of cystathionine gamma lyase (XometC) from Xanthomonas oryzae pv. oryzae in complex with E-site serine, A-site external aldimine structure with serine and A-site external aldimine structure with aminoacrylate intermediates [Xanthomonas oryzae pv. oryzae KACC 10331],4IY7_C crystal structure of cystathionine gamma lyase (XometC) from Xanthomonas oryzae pv. oryzae in complex with E-site serine, A-site external aldimine structure with serine and A-site external aldimine structure with aminoacrylate intermediates [Xanthomonas oryzae pv. oryzae KACC 10331],4IY7_D crystal structure of cystathionine gamma lyase (XometC) from Xanthomonas oryzae pv. oryzae in complex with E-site serine, A-site external aldimine structure with serine and A-site external aldimine structure with aminoacrylate intermediates [Xanthomonas oryzae pv. oryzae KACC 10331],4IYO_A Crystal structure of cystathionine gamma lyase from Xanthomonas oryzae pv. oryzae (XometC) in complex with E-site serine, A-site serine, A-site external aldimine structure with aminoacrylate and A-site iminopropionate intermediates [Xanthomonas oryzae pv. oryzae KACC 10331] |
|
| 9.91e-40 | 80 | 404 | 61 | 395 | Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae [Xanthomonas oryzae pv. oryzae],3E6G_B Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae [Xanthomonas oryzae pv. oryzae],3E6G_C Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae [Xanthomonas oryzae pv. oryzae],3E6G_D Crystal structure of XometC, a cystathionine c-lyase-like protein from Xanthomonas oryzae pv.oryzae [Xanthomonas oryzae pv. oryzae] |
|
| 1.56e-39 | 80 | 406 | 56 | 392 | Chain A, Cystathionine gamma-lyase [Stenotrophomonas maltophilia R551-3],6K1O_B Chain B, Cystathionine gamma-lyase [Stenotrophomonas maltophilia R551-3],6K1O_C Chain C, Cystathionine gamma-lyase [Stenotrophomonas maltophilia R551-3],6K1O_D Chain D, Cystathionine gamma-lyase [Stenotrophomonas maltophilia R551-3] |
|
| 1.56e-39 | 80 | 406 | 56 | 392 | Chain A, Cystathionine gamma-lyase [Stenotrophomonas maltophilia R551-3],6K1L_B Chain B, Cystathionine gamma-lyase [Stenotrophomonas maltophilia R551-3],6K1L_C Chain C, Cystathionine gamma-lyase [Stenotrophomonas maltophilia R551-3],6K1L_D Chain D, Cystathionine gamma-lyase [Stenotrophomonas maltophilia R551-3] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.37e-106 | 16 | 410 | 2 | 376 | Uncharacterized trans-sulfuration enzyme YHR112C OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=YHR112C PE=1 SV=1 |
|
| 1.45e-89 | 17 | 408 | 15 | 397 | Uncharacterized trans-sulfuration enzyme C23A1.14c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC23A1.14c PE=2 SV=1 |
|
| 1.26e-41 | 19 | 410 | 10 | 394 | Cystathionine gamma-lyase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CYS3 PE=1 SV=2 |
|
| 1.77e-39 | 80 | 408 | 44 | 379 | Cystathionine gamma-lyase OS=Bacillus subtilis (strain 168) OX=224308 GN=mccB PE=1 SV=1 |
|
| 5.65e-39 | 80 | 408 | 49 | 384 | Cystathionine beta-lyase MetC OS=Bacillus subtilis (strain 168) OX=224308 GN=metC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
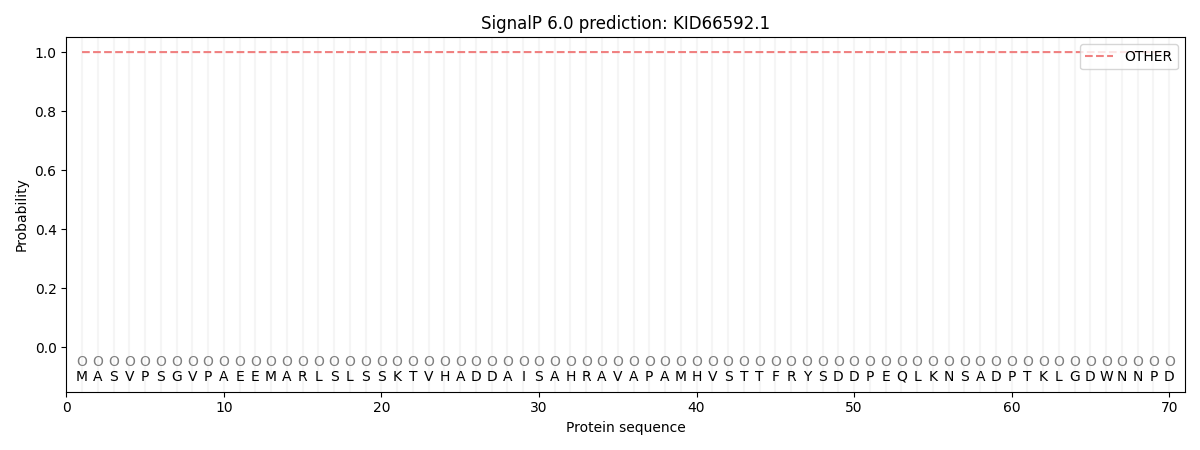
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000056 | 0.000003 |