You are browsing environment: FUNGIDB
CAZyme Information: KID64653.1
You are here: Home > Sequence: KID64653.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
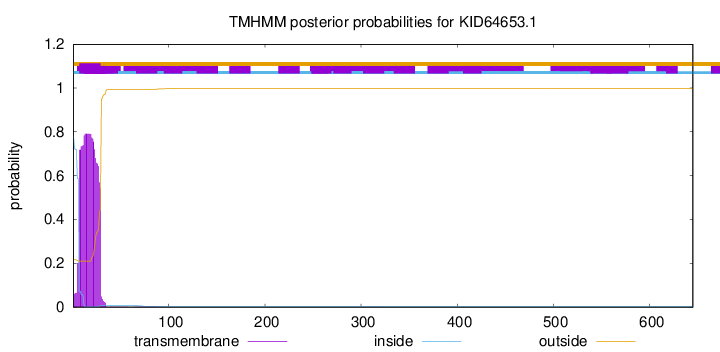
TMHMM annotations
Basic Information help
| Species | Metarhizium anisopliae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Clavicipitaceae; Metarhizium; Metarhizium anisopliae | |||||||||||
| CAZyme ID | KID64653.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | Glycoside hydrolase, family 35 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.23:2 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 41 | 362 | 2e-95 | 0.993485342019544 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396048 | Glyco_hydro_35 | 1.29e-106 | 40 | 362 | 1 | 316 | Glycosyl hydrolases family 35. |
| 166698 | PLN03059 | 2.82e-56 | 19 | 589 | 15 | 659 | beta-galactosidase; Provisional |
| 224786 | GanA | 3.65e-38 | 34 | 641 | 1 | 610 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| 396834 | Glyco_hydro_42 | 1.16e-08 | 57 | 190 | 4 | 139 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| 404274 | BetaGal_dom4_5 | 2.94e-04 | 546 | 616 | 35 | 104 | Beta-galactosidase jelly roll domain. This domain is found in beta galactosidase enzymes. It has a jelly roll fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 645 | 1 | 643 | |
| 3.58e-301 | 1 | 643 | 1 | 634 | |
| 5.97e-289 | 19 | 643 | 20 | 648 | |
| 5.19e-265 | 13 | 597 | 3 | 588 | |
| 1.15e-190 | 28 | 643 | 31 | 633 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.25e-118 | 33 | 618 | 17 | 578 | Chain A, Beta-galactosidase [Niallia circulans],4MAD_B Chain B, Beta-galactosidase [Niallia circulans] |
|
| 2.60e-107 | 32 | 618 | 26 | 593 | Galactanase BT0290 [Bacteroides thetaiotaomicron VPI-5482] |
|
| 4.58e-102 | 32 | 617 | 6 | 572 | Crystal structure of a beta-galactosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
|
| 1.47e-100 | 36 | 619 | 13 | 599 | Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_B Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_C Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_D Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THD_A Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_B Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_C Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_D Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens] |
|
| 2.68e-100 | 36 | 619 | 37 | 623 | Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_B Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_C Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WEZ_D Crystal structure of human beta-galactosidase in complex with NOEV [Homo sapiens],3WF0_A Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_B Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_C Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF0_D Crystal structure of human beta-galactosidase in complex with 6S-NBI-DGJ [Homo sapiens],3WF1_A Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_B Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_C Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF1_D Crystal structure of human beta-galactosidase in complex with 6S-NBI-GJ [Homo sapiens],3WF2_A Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_B Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_C Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens],3WF2_D Crystal structure of human beta-galactosidase in complex with NBT-DGJ [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.54e-108 | 20 | 599 | 13 | 573 | Beta-galactosidase OS=Xanthomonas manihotis OX=43353 GN=bga PE=1 SV=1 |
|
| 9.83e-104 | 40 | 617 | 53 | 613 | Beta-galactosidase-1-like protein 2 OS=Mus musculus OX=10090 GN=Glb1l2 PE=1 SV=1 |
|
| 3.17e-103 | 19 | 619 | 18 | 623 | Beta-galactosidase OS=Canis lupus familiaris OX=9615 GN=GLB1 PE=1 SV=3 |
|
| 4.89e-102 | 19 | 640 | 18 | 644 | Beta-galactosidase OS=Felis catus OX=9685 GN=GLB1 PE=2 SV=1 |
|
| 7.26e-101 | 19 | 619 | 17 | 622 | Beta-galactosidase OS=Macaca fascicularis OX=9541 GN=GLB1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
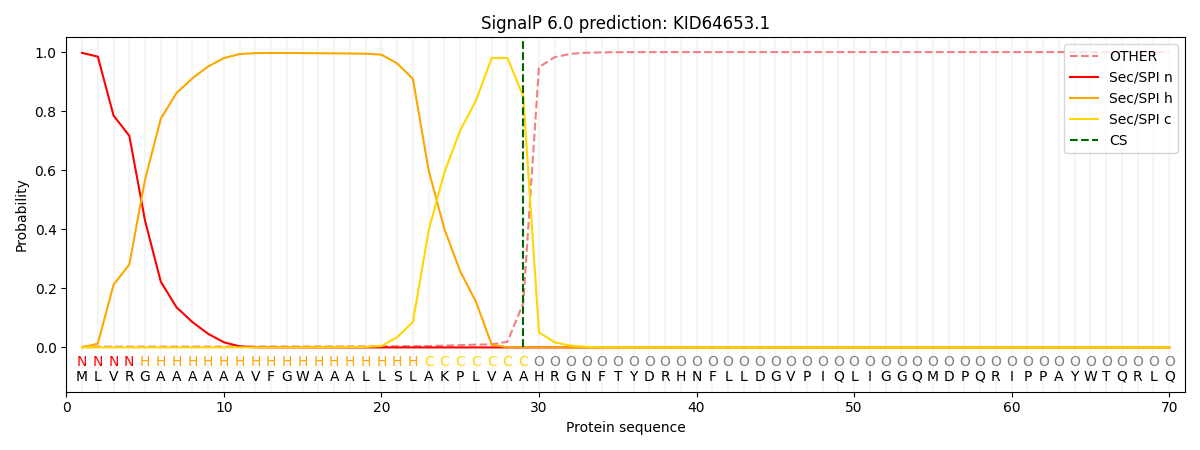
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.004735 | 0.995246 | CS pos: 29-30. Pr: 0.8505 |