You are browsing environment: FUNGIDB
CAZyme Information: KGB76365.1
Basic Information
help
| Species |
Cryptococcus gattii VGII
|
| Lineage |
Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus gattii VGII
|
| CAZyme ID |
KGB76365.1
|
| CAZy Family |
GH154 |
| CAZyme Description |
endoglucanase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 329 |
CP025765|CGC1 |
33812.14 |
6.9993 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_CgattiiVGIIR265 |
6705 |
294750 |
281 |
6424
|
|
| Gene Location |
No EC number prediction in KGB76365.1.
| Family |
Start |
End |
Evalue |
family coverage |
| AA11 |
37 |
209 |
2.7e-58 |
0.9424083769633508 |
KGB76365.1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.30e-202 |
1 |
329 |
1 |
329 |
| 6.70e-151 |
1 |
323 |
1 |
359 |
| 6.30e-149 |
1 |
234 |
1 |
234 |
| 2.30e-66 |
12 |
218 |
14 |
219 |
| 1.07e-64 |
9 |
219 |
11 |
220 |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 8.35e-30 |
42 |
208 |
12 |
203 |
Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Zn [Aspergillus oryzae],4MAI_A Structure of Aspergillus oryzae AA11 Lytic Polysaccharide Monooxygenase with Cu(I) [Aspergillus oryzae] |
KGB76365.1 has no Swissprot hit.
This protein is predicted as SP
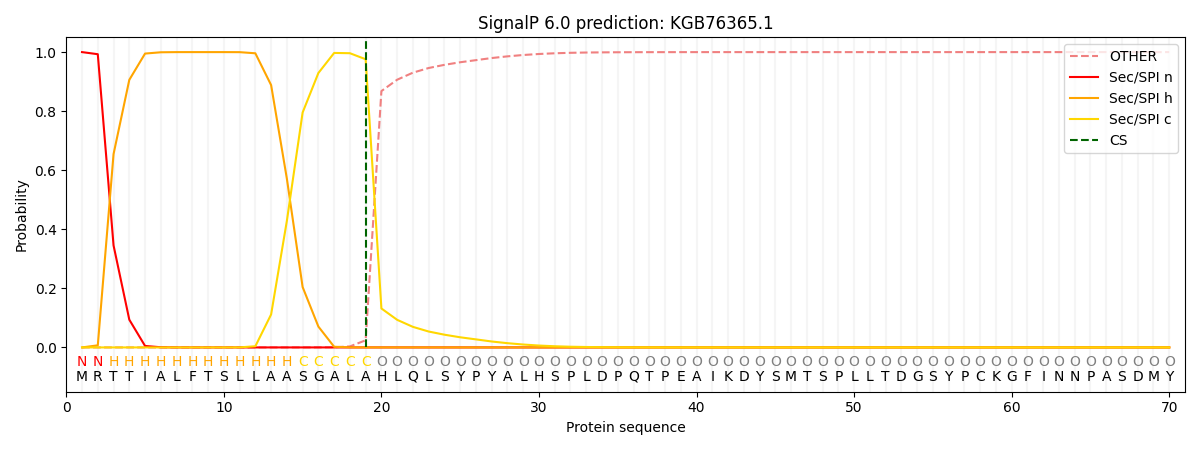
| Other |
SP_Sec_SPI |
CS Position |
| 0.000418 |
0.999560 |
CS pos: 19-20. Pr: 0.9758 |
There is no transmembrane helices in KGB76365.1.

