You are browsing environment: FUNGIDB
CAZyme Information: KGB76302.2
You are here: Home > Sequence: KGB76302.2
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Cryptococcus gattii VGII | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus gattii VGII | |||||||||||
| CAZyme ID | KGB76302.2 | |||||||||||
| CAZy Family | GH13|GH13 | |||||||||||
| CAZyme Description | glucan endo-1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.59:3 |
|---|
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397634 | Glyco_hydro_71 | 1.61e-146 | 394 | 772 | 3 | 372 | Glycosyl hydrolase family 71. Family of alpha-1,3-glucanases. |
| 211418 | GH71 | 2.01e-92 | 386 | 678 | 1 | 283 | Glycoside hydrolase family 71. This family of glycoside hydrolases 71 (following the CAZY nomenclature) function as alpha-1,3-glucanases (mutanases, EC 3.2.1.59). They appear to have an endo-hydrolytic mode of enzymatic activity and bacterial members are investigated as candidates for the development of dental caries treatments.The member from fission yeast, endo-alpha-1,3-glucanase Agn1p, plays a vital role in daughter cell separation, while Agn2p has been associated with endolysis of the ascus wall. |
| 396406 | WSC | 1.39e-25 | 148 | 226 | 1 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 396406 | WSC | 3.31e-25 | 50 | 128 | 1 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
| 396406 | WSC | 3.07e-24 | 259 | 337 | 1 | 80 | WSC domain. This domain may be involved in carbohydrate binding. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 1 | 797 | 1 | 797 | |
| 0.0 | 1 | 783 | 1 | 783 | |
| 0.0 | 1 | 797 | 1 | 800 | |
| 0.0 | 1 | 797 | 1 | 800 | |
| 0.0 | 1 | 783 | 1 | 786 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.69e-14 | 40 | 132 | 83 | 175 | Wnt modulator Kremen in complex with DKK1 (CRD2) and LRP6 (PE3PE4) [Homo sapiens] |
|
| 1.30e-13 | 40 | 132 | 92 | 184 | Chain E, KRM1 [Homo sapiens],7BZU_E Chain E, KRM1 [Homo sapiens] |
|
| 1.33e-13 | 40 | 132 | 95 | 187 | Chain E, Kremen protein 1 [Homo sapiens] |
|
| 1.60e-13 | 40 | 132 | 123 | 215 | Wnt modulator Kremen crystal form I at 1.90A [Homo sapiens],5FWT_A Wnt modulator Kremen crystal form I at 2.10A [Homo sapiens],5FWU_A Wnt modulator Kremen crystal form II at 2.8A [Homo sapiens],5FWV_A Wnt modulator Kremen crystal form III at 3.2A [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.19e-72 | 381 | 749 | 13 | 376 | Glucan endo-1,3-alpha-glucosidase agn1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=agn1 PE=1 SV=2 |
|
| 3.09e-48 | 380 | 758 | 1 | 386 | Ascus wall endo-1,3-alpha-glucanase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=agn2 PE=1 SV=2 |
|
| 1.92e-47 | 387 | 796 | 33 | 429 | Mutanase Pc12g07500 OS=Penicillium rubens (strain ATCC 28089 / DSM 1075 / NRRL 1951 / Wisconsin 54-1255) OX=500485 GN=PCH_Pc12g07500 PE=1 SV=1 |
|
| 5.79e-38 | 40 | 336 | 56 | 393 | WSC domain-containing protein ARB_07867 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07867 PE=1 SV=1 |
|
| 5.41e-30 | 43 | 234 | 521 | 716 | WSC domain-containing protein ARB_07870 OS=Arthroderma benhamiae (strain ATCC MYA-4681 / CBS 112371) OX=663331 GN=ARB_07870 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
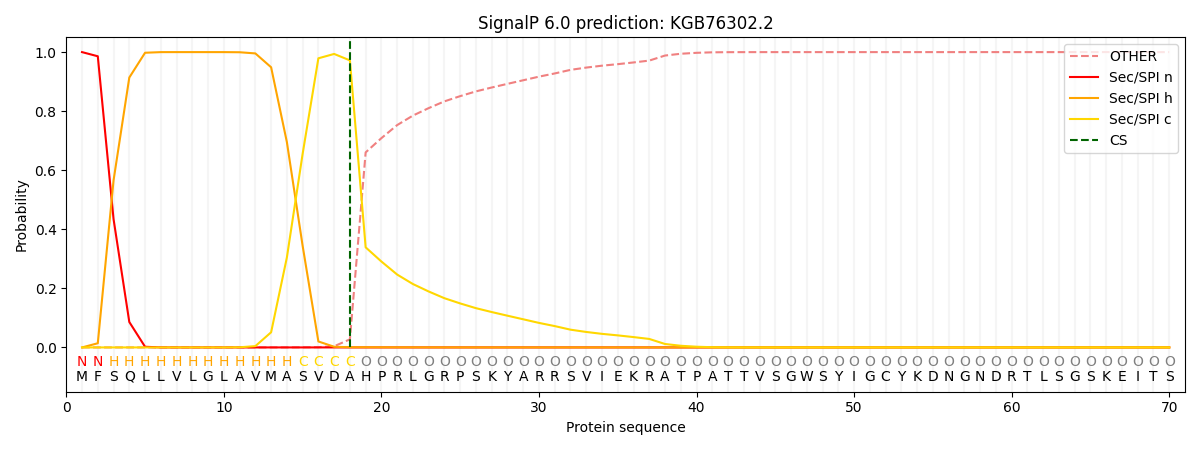
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000482 | 0.999490 | CS pos: 18-19. Pr: 0.9720 |
