You are browsing environment: FUNGIDB
KGB74481.2
Basic Information
help
Species
Cryptococcus gattii VGII
Lineage
Arthropoda; Insecta; ; Eriococcidae; Cryptococcus; Cryptococcus gattii VGII
CAZyme ID
KGB74481.2
CAZy Family
AA14
CAZyme Description
conserved hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
438
45772.42
4.9410
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_CgattiiVGIIR265
6705
294750
281
6424
Gene Location
No EC number prediction in KGB74481.2.
Family
Start
End
Evalue
family coverage
GH128
211
430
1.9e-52
0.96875
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
371727
Glyco_hydro_cc
2.24e-49
201
430
3
235
Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
4.14e-79
181
438
11
270
9.82e-79
192
438
6
254
5.53e-78
192
438
6
254
9.27e-78
181
438
11
270
4.92e-76
192
438
6
254
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
2.25e-99
193
438
198
444
Chain A, Glyco_hydro_cc domain-containing protein [Cryptococcus neoformans]
more
7.36e-80
192
438
28
276
Chain A, Glyco_hydro_cc domain-containing protein [Trichoderma gamsii]
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
2.15e-11
200
432
304
530
Alkali-sensitive linkage protein 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=asl1 PE=1 SV=1
more
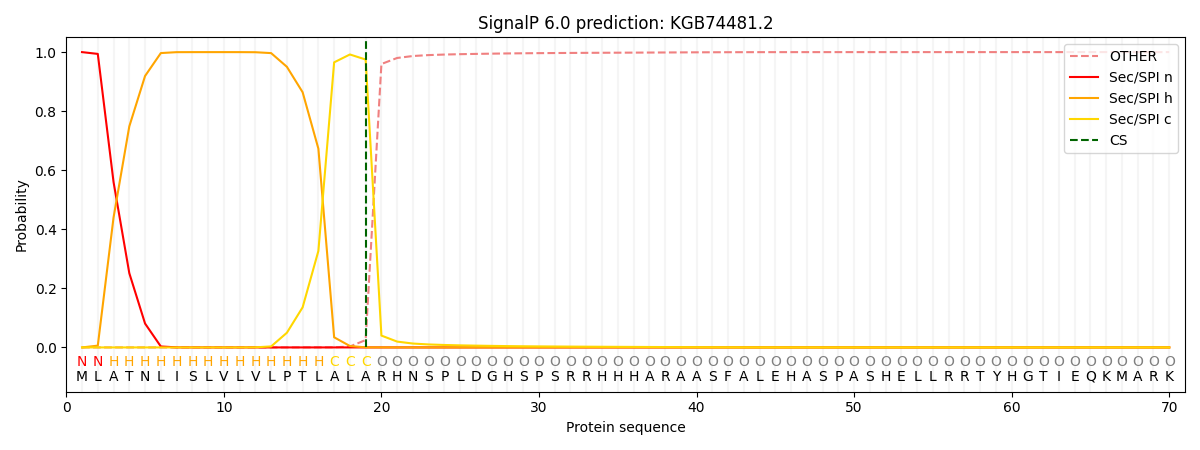
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.000462
0.999500
CS pos: 19-20. Pr: 0.9749
There is no transmembrane helices in KGB74481.2.