You are browsing environment: FUNGIDB
CAZyme Information: KFA54598.1
You are here: Home > Sequence: KFA54598.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stachybotrys chartarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Stachybotryaceae; Stachybotrys; Stachybotrys chartarum | |||||||||||
| CAZyme ID | KFA54598.1 | |||||||||||
| CAZy Family | GH72 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.8:61 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH11 | 52 | 227 | 7.7e-73 | 0.9887005649717514 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395367 | Glyco_hydro_11 | 9.52e-103 | 52 | 225 | 1 | 174 | Glycosyl hydrolases family 11. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.33e-158 | 1 | 231 | 1 | 231 | |
| 2.77e-151 | 1 | 231 | 1 | 231 | |
| 1.99e-139 | 1 | 231 | 1 | 229 | |
| 1.99e-139 | 1 | 231 | 1 | 229 | |
| 8.37e-139 | 1 | 232 | 1 | 231 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.51e-86 | 43 | 229 | 3 | 190 | Chain A, Endo-1,4-beta-xylanase [Thermochaetoides thermophila],1H1A_B Chain B, Endo-1,4-beta-xylanase [Thermochaetoides thermophila] |
|
| 5.34e-86 | 43 | 229 | 3 | 190 | Chain A, endoxylanase 11A [Thermochaetoides thermophila],1XNK_B Chain B, endoxylanase 11A [Thermochaetoides thermophila] |
|
| 8.46e-84 | 43 | 231 | 2 | 188 | 3D Structure of a thermophilic family GH11 xylanase from Thermobifida fusca [Thermobifida fusca] |
|
| 1.49e-83 | 43 | 227 | 4 | 186 | Thermophilic b-1,4-xylanase from Nonomuraea flexuosa [Thermopolyspora flexuosa] |
|
| 8.01e-83 | 43 | 227 | 6 | 188 | Chain A, Endo-1,4-beta-xylanase [Actinomycetia bacterium],7EO6_B Chain B, Endo-1,4-beta-xylanase [Actinomycetia bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.02e-130 | 1 | 231 | 1 | 230 | Endo-1,4-beta-xylanase A OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=XYLA PE=1 SV=1 |
|
| 4.68e-129 | 1 | 232 | 1 | 233 | Endo-1,4-beta-xylanase G OS=Verticillium dahliae OX=27337 GN=xynG PE=1 SV=1 |
|
| 2.34e-119 | 1 | 231 | 1 | 226 | Endo-1,4-beta-xylanase 2 OS=Humicola grisea var. thermoidea OX=5528 GN=xyn2 PE=1 SV=1 |
|
| 4.71e-119 | 1 | 231 | 1 | 226 | Endo-1,4-beta-xylanase 1 OS=Humicola insolens OX=34413 GN=XYL1 PE=2 SV=1 |
|
| 3.82e-105 | 1 | 229 | 1 | 230 | Probable endo-1,4-beta-xylanase A OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=xlnA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
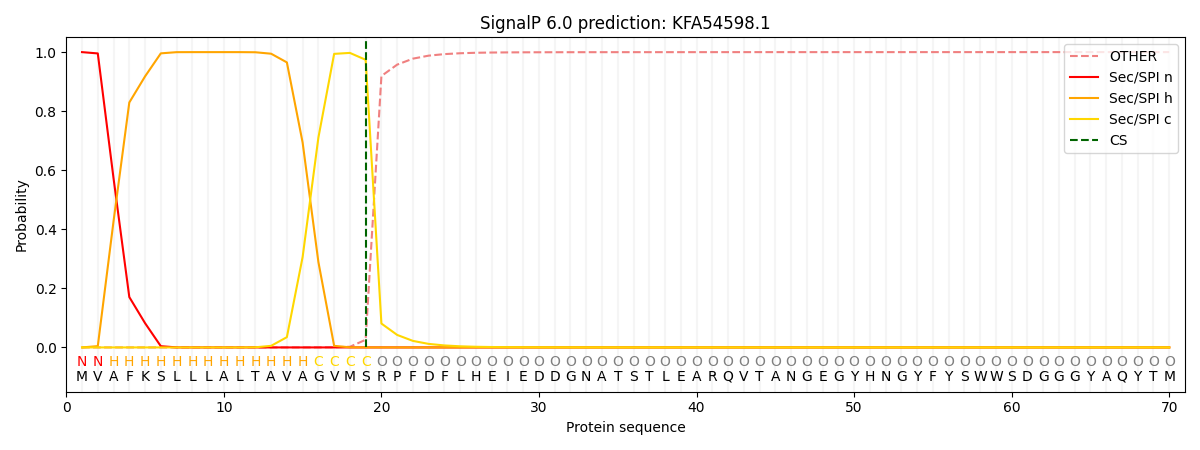
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000311 | 0.999616 | CS pos: 19-20. Pr: 0.9740 |
