You are browsing environment: FUNGIDB
CAZyme Information: KFA53960.1
You are here: Home > Sequence: KFA53960.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stachybotrys chartarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Stachybotryaceae; Stachybotrys; Stachybotrys chartarum | |||||||||||
| CAZyme ID | KFA53960.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 264048; End:272465 Strand: + | |||||||||||
Full Sequence Download help
| MTGLGFSTAA AYPGFGALNG YWGQFGSESL RQYCDTGIEY VTLSFVNQAP EHSASNYPGT | 60 |
| NFAGHCWANT YTAPNGASSK LLSECRTIKN DIPYCREKGV KVLLSIGGEY SEEYSNYRVT | 120 |
| STANGEYFAN FLYKAFGPYD PSWTGPRPFD LDANTHTQVD GFDFDIEADF DNEPYIRMAE | 180 |
| KFRELDSSMF LTAAPQCPTL DPYFHLKDLI QRAPLDALFV QFYNNAVCDA IRDPGLAGDG | 240 |
| FNYNAWANLL AQSDKSKNAK LFIGLLASPA ASLTGSGFIS AQAMKDLVCQ YKDSPSFGGI | 300 |
| SLWDLTRGAA EISNGKNYYQ HALDALKYGC APVPTSTSRA ITSTTSTRTS TSTSTRQPTT | 360 |
| TTTTIRSSSS SSVLTSTSTT SSSIRPTTTS TTVRSSSTTS PSSTLISTTS ARTSTVTSSS | 420 |
| SSSSSSSSSS SSSISSSSTS SSSSSSSIMS SSSSSSSTSS SSSSSSSSTT TTSSGIIIET | 480 |
| PSVSDTSLIS DVFTSVTSAA STSTLSASSS MTTTSSGIII ETPSVSDTSL ISDVFTSVTS | 540 |
| AASTTSAETV SASVSSAVSV PTTTTVVTSL PVSFTSTRTW SNSTMTTSSA PGTSDVSVSA | 600 |
| PTTTTVMTSL PGSLTSTRSW SNTTFTTSDA GSLTTRVASL TTSAVSLTTS TVYTTKVQTV | 660 |
| TKCPPYVVDC PEGGYVTTVT VPWYTTVCPV TETPKPTGAL TTSTVYATKV YTVTKCPPGV | 720 |
| ANCPTGHATT ETVPVYTTVC PVGETVTVPL TTSTAYTTAT YTITACPEGV VDCPTGHMTT | 780 |
| VTEPAYTTIG PIEETSVVPE PTGDAETQTY TLTYPHGGAE TSTVVVPGGG DAESSTRPGV | 840 |
| AVPTDQWVNP IPSGTPGLPD VPNVPASAPS LAVGLGGLLA LVAVQVFALF SDCVFDFHIS | 900 |
| GLLDAKNPDS LHKIACFEMR LFSGVVKLQL SKALVQRIAV HEEIRNFREE SHCQASLFQD | 960 |
| LEQPEDQTKA LMQKWTTCQD LGNAAPEPIK KLALGKHWTP PLFDIEMSRT LANHLAICPV | 1020 |
| SNTSQHKRSK SAAALALLRR KDTREDDSGS EDARSAQPPS SASKPGNSSM AHKSSARSSN | 1080 |
| SKLTTGSALS RAPSHQPSPA AKPTPAPDRG VTLEQSVKKF RIVEALRNGD TAYISRAIRD | 1140 |
| TAENGPRTSI SSIATTSASA ALEDTTLLHL AIQCAEFPVI EYVLSDGLGS IDFNARDKDG | 1200 |
| NTPLHVAATQ GRAPVVKLLL EQKDINDAIS NSQGKLPIDL ARNPEIFQLL QLSRSLFTEA | 1260 |
| KIDQVQGLIA SRKYDTLAEV LEESRVKTVL DINSPEFASE PVTVQTGGTL LHEAVRKRDT | 1320 |
| KLIEVLLLHG ADPFCRDRKG KLPQSITSDD NIKSILKKSP AAVAAQRGIQ EKAVLGHAAS | 1380 |
| QGAATTDPLA GREAREMKGY LKKWTNYRKG YQLRWFVLED GVLSYYKHQD DSGSACRGAI | 1440 |
| NMRIAQLHMT ADEKTKFEII GKSSVKYTLK ANHEVEAKRW FWALNNSIQW TKDQIKEEQR | 1500 |
| QAARSAEMLK TAKAEQSGSL SLQESHSESA SLSDLRRNSS HYGSRSIGGK GPTQKPEFGT | 1560 |
| STVGSIDEDD EFADAATDAG ASRAGHGHLI PGDPDDDDDD YGDGSLMEQP SATKDALNIT | 1620 |
| AQSAKLQLET MSHVHNALMG ELTQSPSTPL SDTKVTQALS TYDAAIRSLT GLVGDLLRIS | 1680 |
| KDRDAYWQYR LDRESNMRRM WEESMAKVAR EHEVLEARVG QAELKRKQTK RALREVMESG | 1740 |
| FTATDVPDSQ PGVAEDEDGQ SRSPTVARRA TIGQIVDLSE SESEDEEEFF DAVDAGEVEV | 1800 |
| SVMPPEDGAK QEADIVVTGG IDLSSSFKGY ENGIRHRLKM DADDRPKISL WGILKSMIGK | 1860 |
| DMTRMTLPVS FNEPTSLLYR CGEDMEYVDL LDLAAERTDS IERLIYVAAF AASEYASTID | 1920 |
| RVAKPFNPLL GETFEYVRPD KNYRFFIEQV SHHPPIGAAW AESPHWTYWG ESHVKSKFYG | 1980 |
| KSFDINPLGT WFLKLRPKAG GKEDLYTWKK VTSSVVGIIT GNPTVDNYGL MEIKNWTTGE | 2040 |
| VAQLDFKPRG WKASSAYQIS GKVMDETGRV RVSMGGRWNS KLYARLTPGY EAALDEPAKD | 2100 |
| SGPVHRGNIT DPTRAYLVWQ ANPRPAGIPF NLTPFVLTFN HIDEKLRPWI APTDSRLRPD | 2160 |
| QRAMEDGEYD FAATEKNRLE QNQRARRQDR DARGEAFVPA WFYKTRCEIT GEEYWQFNGK | 2220 |
| YWEQREKAGP NGDADAWAGL EPIYEDYEAK NE | 2252 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 19 | 276 | 2.5e-18 | 0.6722972972972973 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 395990 | Oxysterol_BP | 0.0 | 1849 | 2223 | 1 | 362 | Oxysterol-binding protein. |
| 119356 | GH18_hevamine_XipI_class_III | 4.29e-92 | 16 | 324 | 1 | 280 | This conserved domain family includes xylanase inhibitor Xip-I, and the class III plant chitinases such as hevamine, concanavalin B, and PPL2, all of which have a glycosyl hydrolase family 18 (GH18) domain. Hevamine is a class III endochitinase that hydrolyzes the linear polysaccharide chains of chitin and peptidoglycan and is important for defense against pathogenic bacteria and fungi. PPL2 (Parkia platycephala lectin 2) is a class III chitinase from Parkia platycephala seeds that hydrolyzes beta(1-4) glycosidic bonds linking 2-acetoamido-2-deoxy-beta-D-glucopyranose units in chitin. |
| 241446 | PH_Osh1p_Osh2p_yeast | 1.34e-58 | 1394 | 1494 | 1 | 103 | Yeast oxysterol binding protein homologs 1 and 2 Pleckstrin homology (PH) domain. Yeast Osh1p is proposed to function in postsynthetic sterol regulation, piecemeal microautophagy of the nucleus, and cell polarity establishment. Yeast Osh2p is proposed to function in sterol metabolism and cell polarity establishment. Both Osh1p and Osh2p contain 3 N-terminal ankyrin repeats, a PH domain, a FFAT motif (two phenylalanines in an acidic tract), and a C-terminal OSBP-related domain. OSBP andOsh1p PH domains specifically localize to the Golgi apparatus in a PtdIns4P-dependent manner. Oxysterol binding proteins are a multigene family that is conserved in yeast, flies, worms, mammals and plants. In general OSBPs and ORPs have been found to be involved in the transport and metabolism of cholesterol and related lipids in eukaryotes. They all contain a C-terminal oxysterol binding domain, and most contain an N-terminal PH domain. OSBP PH domains bind to membrane phosphoinositides and thus likely play an important role in intracellular targeting. They are members of the oxysterol binding protein (OSBP) family which includes OSBP, OSBP-related proteins (ORP), Goodpasture antigen binding protein (GPBP), and Four phosphate adaptor protein 1 (FAPP1). They have a wide range of purported functions including sterol transport, cell cycle control, pollen development and vessicle transport from Golgi recognize both PI lipids and ARF proteins. PH domains have diverse functions, but in general are involved in targeting proteins to the appropriate cellular location or in the interaction with a binding partner. They share little sequence conservation, but all have a common fold, which is electrostatically polarized. Less than 10% of PH domains bind phosphoinositide phosphates (PIPs) with high affinity and specificity. PH domains are distinguished from other PIP-binding domains by their specific high-affinity binding to PIPs with two vicinal phosphate groups: PtdIns(3,4)P2, PtdIns(4,5)P2 or PtdIns(3,4,5)P3 which results in targeting some PH domain proteins to the plasma membrane. A few display strong specificity in lipid binding. Any specificity is usually determined by loop regions or insertions in the N-terminus of the domain, which are not conserved across all PH domains. PH domains are found in cellular signaling proteins such as serine/threonine kinase, tyrosine kinases, regulators of G-proteins, endocytotic GTPases, adaptors, as well as cytoskeletal associated molecules and in lipid associated enzymes. |
| 270101 | PH_OSBP_ORP4 | 5.97e-28 | 1397 | 1489 | 1 | 91 | Human Oxysterol binding protein and OSBP-related protein 4 Pleckstrin homology (PH) domain. Human OSBP is proposed to function is sterol-dependent regulation of ERK dephosphorylation and sphingomyelin synthesis as well as modulation of insulin signaling and hepatic lipogenesis. It contains a N-terminal PH domain, a FFAT motif (two phenylalanines in an acidic tract), and a C-terminal OSBP-related domain. OSBPs and Osh1p PH domains specifically localize to the Golgi apparatus in a PtdIns4P-dependent manner. ORP4 is proposed to function in Vimentin-dependent sterol transport and/or signaling. Human ORP4 has 2 forms, a long (ORP4L) and a short (ORP4S). ORP4L contains a N-terminal PH domain, a FFAT motif (two phenylalanines in an acidic tract), and a C-terminal OSBP-related domain. ORP4S is truncated and contains only an OSBP-related domain. Oxysterol binding proteins are a multigene family that is conserved in yeast, flies, worms, mammals and plants. They all contain a C-terminal oxysterol binding domain, and most contain an N-terminal PH domain. OSBP PH domains bind to membrane phosphoinositides and thus likely play an important role in intracellular targeting. They are members of the oxysterol binding protein (OSBP) family which includes OSBP, OSBP-related proteins (ORP), Goodpasture antigen binding protein (GPBP), and Four phosphate adaptor protein 1 (FAPP1). They have a wide range of purported functions including sterol transport, cell cycle control, pollen development and vessicle transport from Golgi recognize both PI lipids and ARF proteins. PH domains have diverse functions, but in general are involved in targeting proteins to the appropriate cellular location or in the interaction with a binding partner. They share little sequence conservation, but all have a common fold, which is electrostatically polarized. Less than 10% of PH domains bind phosphoinositide phosphates (PIPs) with high affinity and specificity. PH domains are distinguished from other PIP-binding domains by their specific high-affinity binding to PIPs with two vicinal phosphate groups: PtdIns(3,4)P2, PtdIns(4,5)P2 or PtdIns(3,4,5)P3 which results in targeting some PH domain proteins to the plasma membrane. A few display strong specificity in lipid binding. Any specificity is usually determined by loop regions or insertions in the N-terminus of the domain, which are not conserved across all PH domains. PH domains are found in cellular signaling proteins such as serine/threonine kinase, tyrosine kinases, regulators of G-proteins, endocytotic GTPases, adaptors, as well as cytoskeletal associated molecules and in lipid associated enzymes. |
| 269951 | PH_FAPP1_FAPP2 | 2.43e-23 | 1397 | 1487 | 1 | 91 | Four phosphate adaptor protein 1 and 2 Pleckstrin homology (PH) domain. Human FAPP1 (also called PLEKHA3/Pleckstrin homology domain-containing, family A member 3) regulates secretory transport from the trans-Golgi network to the plasma membrane. It is recruited through binding of PH domain to phosphatidylinositol 4-phosphate (PtdIns(4)P) and a small GTPase ADP-ribosylation factor 1 (ARF1). These two binding sites have little overlap the FAPP1 PH domain to associate with both ligands simultaneously and independently. FAPP1 has a N-terminal PH domain followed by a short proline-rich region. FAPP1 is a member of the oxysterol binding protein (OSBP) family which includes OSBP, OSBP-related proteins (ORP), and Goodpasture antigen binding protein (GPBP). They have a wide range of purported functions including sterol transport, cell cycle control, pollen development and vessicle transport from Golgi recognize both PI lipids and ARF proteins. FAPP2 (also called PLEKHA8/Pleckstrin homology domain-containing, family A member 8), a member of the Glycolipid lipid transfer protein(GLTP) family has an N-terminal PH domain that targets the TGN and C-terminal GLTP domain. FAPP2 functions to traffic glucosylceramide (GlcCer) which is made in the Golgi. It's interaction with vesicle-associated membrane protein-associated protein (VAP) could be a means of regulation. Some FAPP2s share the FFAT-like motifs found in GLTP. PH domains have diverse functions, but in general are involved in targeting proteins to the appropriate cellular location or in the interaction with a binding partner. They share little sequence conservation, but all have a common fold, which is electrostatically polarized. Less than 10% of PH domains bind phosphoinositide phosphates (PIPs) with high affinity and specificity. PH domains are distinguished from other PIP-binding domains by their specific high-affinity binding to PIPs with two vicinal phosphate groups: PtdIns(3,4)P2, PtdIns(4,5)P2 or PtdIns(3,4,5)P3 which results in targeting some PH domain proteins to the plasma membrane. A few display strong specificity in lipid binding. Any specificity is usually determined by loop regions or insertions in the N-terminus of the domain, which are not conserved across all PH domains. PH domains are found in cellular signaling proteins such as serine/threonine kinase, tyrosine kinases, regulators of G-proteins, endocytotic GTPases, adaptors, as well as cytoskeletal associated molecules and in lipid associated enzymes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKD55908.1|GH18 | 0.0 | 18 | 2246 | 16 | 2188 |
| CCT69979.1|GH18 | 4.43e-122 | 18 | 336 | 16 | 334 |
| QGI96702.1|GH18 | 4.43e-122 | 18 | 336 | 16 | 334 |
| QGI65821.1|GH18 | 4.43e-122 | 18 | 336 | 16 | 334 |
| QGI83062.1|GH18 | 2.04e-121 | 18 | 336 | 16 | 334 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5H2D_A | 1.83e-141 | 1826 | 2233 | 21 | 433 | Crystal structure of Osh1 ORD domain in complex with ergosterol [Kluyveromyces lactis NRRL Y-1140],5WVR_A Crystal structure of Osh1 ORD domain in complex with cholesterol [Kluyveromyces lactis NRRL Y-1140] |
| 7DEI_A | 2.85e-74 | 1824 | 2229 | 2 | 378 | Chain A, Oxysterol-binding protein-related protein 3 [Homo sapiens],7DEI_B Chain B, Oxysterol-binding protein-related protein 3 [Homo sapiens],7DEJ_A Chain A, Oxysterol-binding protein-related protein 3 [Homo sapiens],7DEJ_B Chain B, Oxysterol-binding protein-related protein 3 [Homo sapiens] |
| 7CYZ_A | 9.55e-74 | 1848 | 2229 | 16 | 367 | The structure of human ORP3 OSBP-related domain [Homo sapiens],7CYZ_B The structure of human ORP3 OSBP-related domain [Homo sapiens] |
| 5ZM6_A | 9.82e-73 | 1832 | 2224 | 6 | 419 | Crystal structure of ORP1-ORD in complex with PI(4,5)P2 [Homo sapiens],5ZM6_B Crystal structure of ORP1-ORD in complex with PI(4,5)P2 [Homo sapiens],5ZM7_A Crystal structure of ORP1-ORD in complex with cholesterol at 3.4 A resolution [Homo sapiens] |
| 5ZM5_A | 1.37e-72 | 1845 | 2224 | 12 | 409 | Crystal structure of human ORP1-ORD in complex with cholesterol at 2.6 A resolution [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P35845|OSH1_YEAST | 8.82e-192 | 1167 | 2225 | 55 | 1176 | Oxysterol-binding protein homolog 1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SWH1 PE=1 SV=4 |
| sp|Q12451|OSH2_YEAST | 2.07e-179 | 1167 | 2225 | 59 | 1271 | Oxysterol-binding protein homolog 2 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=OSH2 PE=1 SV=1 |
| sp|O14340|YB35_SCHPO | 1.15e-175 | 1113 | 2244 | 22 | 1310 | Oxysterol-binding protein homolog C2F12.05c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC2F12.05c PE=1 SV=2 |
| sp|Q3B7Z2|OSBP1_MOUSE | 8.49e-87 | 1394 | 2227 | 88 | 795 | Oxysterol-binding protein 1 OS=Mus musculus OX=10090 GN=Osbp PE=1 SV=3 |
| sp|P22059|OSBP1_HUMAN | 2.39e-85 | 1394 | 2227 | 90 | 797 | Oxysterol-binding protein 1 OS=Homo sapiens OX=9606 GN=OSBP PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
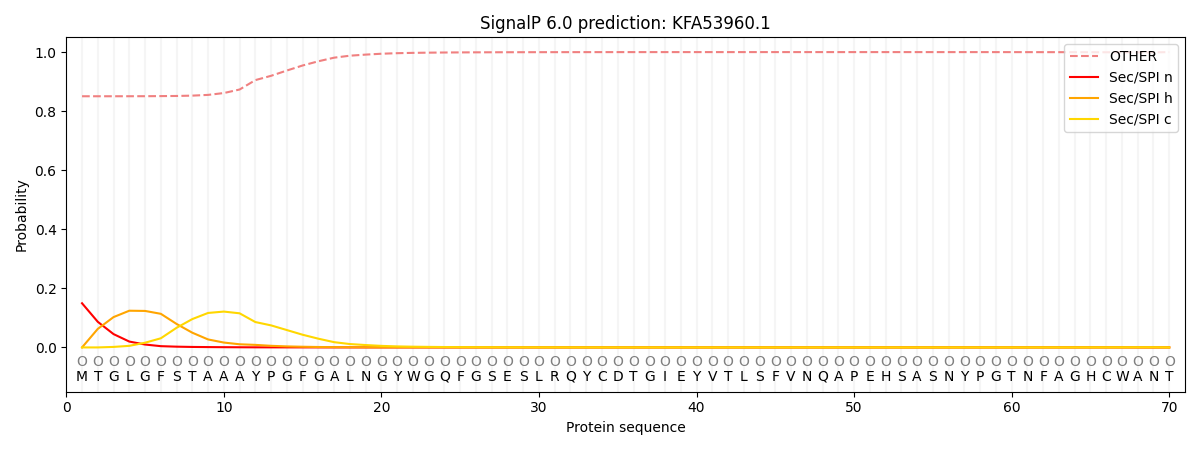
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.859363 | 0.140655 |
