You are browsing environment: FUNGIDB
CAZyme Information: KFA53412.1
You are here: Home > Sequence: KFA53412.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stachybotrys chartarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Stachybotryaceae; Stachybotrys; Stachybotrys chartarum | |||||||||||
| CAZyme ID | KFA53412.1 | |||||||||||
| CAZy Family | GH43 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.258:10 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT58 | 590 | 954 | 3.1e-143 | 0.9945054945054945 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398745 | ALG3 | 0.0 | 590 | 954 | 1 | 358 | ALG3 protein. The formation of N-glycosidic linkages of glycoproteins involves the ordered assembly of the common Glc3Man9GlcNAc2 core-oligosaccharide on the lipid carrier dolichyl pyrophosphate. Whereas early mannosylation steps occur on the cytoplasmic side of the endoplasmic reticulum with GDP-Man as donor, the final reactions from Man5GlcNAc2-PP-Dol to Man9GlcNAc2-PP-Dol on the lumenal side use Dol-P-Man. ALG3 gene encodes the Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase. |
| 239245 | TRX_family | 1.62e-29 | 44 | 136 | 1 | 93 | TRX family; composed of two groups: Group I, which includes proteins that exclusively encode a TRX domain; and Group II, which are composed of fusion proteins of TRX and additional domains. Group I TRX is a small ancient protein that alter the redox state of target proteins via the reversible oxidation of an active site dithiol, present in a CXXC motif, partially exposed at the protein's surface. TRX reduces protein disulfide bonds, resulting in a disulfide bond at its active site. Oxidized TRX is converted to the active form by TRX reductase, using reducing equivalents derived from either NADPH or ferredoxins. By altering their redox state, TRX regulates the functions of at least 30 target proteins, some of which are enzymes and transcription factors. It also plays an important role in the defense against oxidative stress by directly reducing hydrogen peroxide and certain radicals, and by serving as a reductant for peroxiredoxins. At least two major types of functional TRXs have been reported in most organisms; in eukaryotes, they are located in the cytoplasm and the mitochondria. Higher plants contain more types (at least 20 TRX genes have been detected in the genome of Arabidopsis thaliana), two of which (types f amd m) are located in the same compartment, the chloroplast. Also included in the alignment are TRX-like domains which show sequence homology to TRX but do not contain the redox active CXXC motif. Group II proteins, in addition to either a redox active TRX or a TRX-like domain, also contain additional domains, which may or may not possess homology to known proteins. |
| 200072 | thioredoxin | 7.40e-28 | 42 | 138 | 2 | 100 | thioredoxin. Several proteins, such as protein disulfide isomerase, have two or more copies of a domain closely related to thioredoxin. This model is designed to recognize authentic thioredoxin, a small protein that should be hit exactly once by this model. Any protein that hits once with a score greater than the second (per domain) trusted cutoff may be taken as thioredoxin. [Energy metabolism, Electron transport] |
| 173347 | PTZ00051 | 3.67e-25 | 37 | 134 | 2 | 98 | thioredoxin; Provisional |
| 396016 | DAO | 3.29e-23 | 185 | 522 | 7 | 339 | FAD dependent oxidoreductase. This family includes various FAD dependent oxidoreductases: Glycerol-3-phosphate dehydrogenase EC:1.1.99.5, Sarcosine oxidase beta subunit EC:1.5.3.1, D-alanine oxidase EC:1.4.99.1, D-aspartate oxidase EC:1.4.3.1. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 185 | 973 | 10 | 777 | |
| 0.0 | 185 | 973 | 10 | 792 | |
| 0.0 | 185 | 973 | 10 | 792 | |
| 0.0 | 185 | 973 | 10 | 792 | |
| 0.0 | 185 | 973 | 10 | 792 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.78e-114 | 185 | 533 | 31 | 388 | Chain A, D-amino acid oxidase [Rasamsonia emersonii],7CT4_B Chain B, D-amino acid oxidase [Rasamsonia emersonii],7CT4_C Chain C, D-amino acid oxidase [Rasamsonia emersonii],7CT4_D Chain D, D-amino acid oxidase [Rasamsonia emersonii] |
|
| 5.70e-47 | 185 | 523 | 14 | 354 | CRYSTAL STRUCTURE OF D-AMINO ACID OXIDASE IN COMPLEX WITH TWO ANTHRANYLATE MOLECULES [Rhodotorula toruloides],1C0K_A Crystal Structure Analysis Of D-amino Acid Oxidase In Complex With L- Lactate [Rhodotorula toruloides],1C0L_A D-AMINO ACID OXIDASE: STRUCTURE OF SUBSTRATE COMPLEXES AT VERY HIGH RESOLUTION REVEAL THE CHEMICAL REACTTION MECHANISM OF FLAVIN DEHYDROGENATION [Rhodotorula toruloides],1C0P_A D-AMINO ACIC OXIDASE IN COMPLEX WITH D-ALANINE AND A PARTIALLY OCCUPIED BIATOMIC SPECIES [Rhodotorula toruloides] |
|
| 1.61e-23 | 36 | 138 | 1 | 104 | Crystal Structure of Ancestral Thioredoxin Relative to Last Eukaryotes Common Ancestor (LECA) from the Precambrian Period [synthetic construct],2YOI_B Crystal Structure of Ancestral Thioredoxin Relative to Last Eukaryotes Common Ancestor (LECA) from the Precambrian Period [synthetic construct] |
|
| 2.33e-21 | 36 | 138 | 1 | 104 | Crystal Structure of Ancestral Thioredoxin Relative to Last Animal and Fungi Common Ancestor (LAFCA) from the Precambrian Period [synthetic construct] |
|
| 1.17e-20 | 20 | 139 | 27 | 135 | Crystal Structure of Thioredoxin NaTrxh from Nicotiana alata [Nicotiana alata],6X0B_B Crystal Structure of Thioredoxin NaTrxh from Nicotiana alata [Nicotiana alata] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.63e-186 | 561 | 990 | 9 | 440 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=alg-3 PE=3 SV=1 |
|
| 9.52e-149 | 185 | 526 | 10 | 353 | D-amino-acid oxidase OS=Fusarium vanettenii OX=2747968 PE=1 SV=1 |
|
| 6.62e-139 | 582 | 986 | 8 | 405 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=alg3 PE=3 SV=1 |
|
| 2.03e-131 | 590 | 962 | 30 | 393 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=alg3 PE=3 SV=1 |
|
| 7.64e-128 | 581 | 962 | 15 | 385 | Dol-P-Man:Man(5)GlcNAc(2)-PP-Dol alpha-1,3-mannosyltransferase OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=alg3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000042 | 0.000000 |
TMHMM Annotations download full data without filtering help
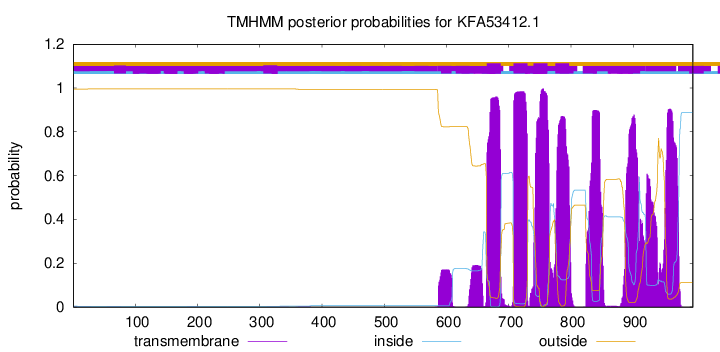
| Start | End |
|---|---|
| 665 | 687 |
| 708 | 730 |
| 740 | 762 |
| 775 | 797 |
