You are browsing environment: FUNGIDB
CAZyme Information: KFA51356.1
You are here: Home > Sequence: KFA51356.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stachybotrys chartarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Stachybotryaceae; Stachybotrys; Stachybotrys chartarum | |||||||||||
| CAZyme ID | KFA51356.1 | |||||||||||
| CAZy Family | GH162 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.1.3.13:12 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 167 | 757 | 1.6e-265 | 0.9898305084745763 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 4.08e-77 | 167 | 759 | 11 | 540 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 235000 | PRK02106 | 2.15e-76 | 167 | 752 | 9 | 531 | choline dehydrogenase; Validated |
| 366272 | GMC_oxred_N | 6.37e-41 | 236 | 475 | 16 | 216 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
| 398739 | GMC_oxred_C | 6.01e-30 | 584 | 748 | 1 | 143 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 381213 | lipocalin_Blc-like | 8.43e-25 | 46 | 169 | 3 | 124 | bacterial lipocalin Blc, Arabidopsis thaliana temperature-induced lipocalin-1, and similar proteins. Escherichia coli bacterial lipocalin (Blc, also known as YjeL) is an outer membrane lipoprotein involved in the storage or transport of lipids necessary for membrane maintenance under stressful conditions. Blc has a binding preference for lysophospholipids. This group includes eukaryotic lipocalins such as Arabidopsis thaliana temperature-induced lipocalin-1 (TIL) which is involved in thermotolerance, oxidative, salt, drought and high light stress tolerance, and is needed for seed longevity by ensuring polyunsaturated lipids integrity. This group belongs to the lipocalin/cytosolic fatty-acid binding protein family which have a large beta-barrel ligand-binding cavity. Lipocalins are mainly low molecular weight extracellular proteins that bind principally small hydrophobic ligands, and form covalent or non-covalent complexes with soluble macromolecules, as well as membrane bound-receptors. They participate in processes such as ligand transport, modulation of cell growth and metabolism, regulation of immune response, smell reception, tissue development and animal behavior. Cytosolic fatty-acid binding proteins, also bind hydrophobic ligands in a non-covalent, reversible manner, and have been implicated in intracellular uptake, transport and storage of hydrophobic ligands, regulation of lipid metabolism and sequestration of excess toxic fatty acids, as well as in signaling, gene expression, inflammation, cell growth and proliferation, and cancer development. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.29e-290 | 167 | 756 | 17 | 603 | |
| 1.24e-283 | 167 | 758 | 17 | 604 | |
| 1.79e-279 | 167 | 758 | 17 | 604 | |
| 1.79e-279 | 167 | 758 | 17 | 604 | |
| 1.79e-279 | 167 | 758 | 17 | 604 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.22e-148 | 167 | 759 | 10 | 627 | Chain A, Alcohol oxidase [Phanerodontia chrysosporium],6H3G_B Chain B, Alcohol oxidase [Phanerodontia chrysosporium],6H3G_C Chain C, Alcohol oxidase [Phanerodontia chrysosporium],6H3G_D Chain D, Alcohol oxidase [Phanerodontia chrysosporium],6H3G_E Chain E, Alcohol oxidase [Phanerodontia chrysosporium],6H3G_F Chain F, Alcohol oxidase [Phanerodontia chrysosporium],6H3G_G Chain G, Alcohol oxidase [Phanerodontia chrysosporium],6H3G_H Chain H, Alcohol oxidase [Phanerodontia chrysosporium] |
|
| 5.04e-147 | 167 | 759 | 10 | 627 | Chain A, Alcohol oxidase [Phanerodontia chrysosporium],6H3O_B Chain B, Alcohol oxidase [Phanerodontia chrysosporium],6H3O_C Chain C, Alcohol oxidase [Phanerodontia chrysosporium],6H3O_D Chain D, Alcohol oxidase [Phanerodontia chrysosporium],6H3O_E Chain E, Alcohol oxidase [Phanerodontia chrysosporium],6H3O_F Chain F, Alcohol oxidase [Phanerodontia chrysosporium],6H3O_G Chain G, Alcohol oxidase [Phanerodontia chrysosporium],6H3O_H Chain H, Alcohol oxidase [Phanerodontia chrysosporium] |
|
| 1.75e-137 | 171 | 760 | 14 | 640 | Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_B Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_C Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_D Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_E Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_F Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_G Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5HSA_H Alcohol Oxidase AOX1 from Pichia Pastoris [Komagataella phaffii CBS 7435],5I68_A Chain A, Alcohol oxidase 1 [Komagataella pastoris] |
|
| 6.60e-43 | 167 | 752 | 5 | 561 | Crystal structure of aryl-alcohol oxidase from Pleurotus eryngii in complex with p-anisic acid [Pleurotus eryngii] |
|
| 3.06e-42 | 167 | 752 | 6 | 562 | Crystal structure of aryl-alcohol-oxidase from Pleurotus eryingii [Pleurotus eryngii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.14e-141 | 167 | 761 | 10 | 641 | Alcohol oxidase OS=Candida boidinii OX=5477 GN=AOD1 PE=1 SV=1 |
|
| 9.02e-137 | 171 | 760 | 14 | 640 | Alcohol oxidase 1 OS=Komagataella phaffii (strain GS115 / ATCC 20864) OX=644223 GN=AOX1 PE=1 SV=2 |
|
| 9.02e-137 | 171 | 760 | 14 | 640 | Alcohol oxidase 1 OS=Komagataella phaffii (strain ATCC 76273 / CBS 7435 / CECT 11047 / NRRL Y-11430 / Wegner 21-1) OX=981350 GN=AOX1 PE=1 SV=1 |
|
| 2.59e-136 | 167 | 761 | 10 | 642 | Alcohol oxidase OS=Pichia angusta OX=870730 GN=MOX PE=1 SV=1 |
|
| 3.55e-136 | 171 | 760 | 14 | 640 | Alcohol oxidase 2 OS=Komagataella phaffii (strain GS115 / ATCC 20864) OX=644223 GN=AOX2 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
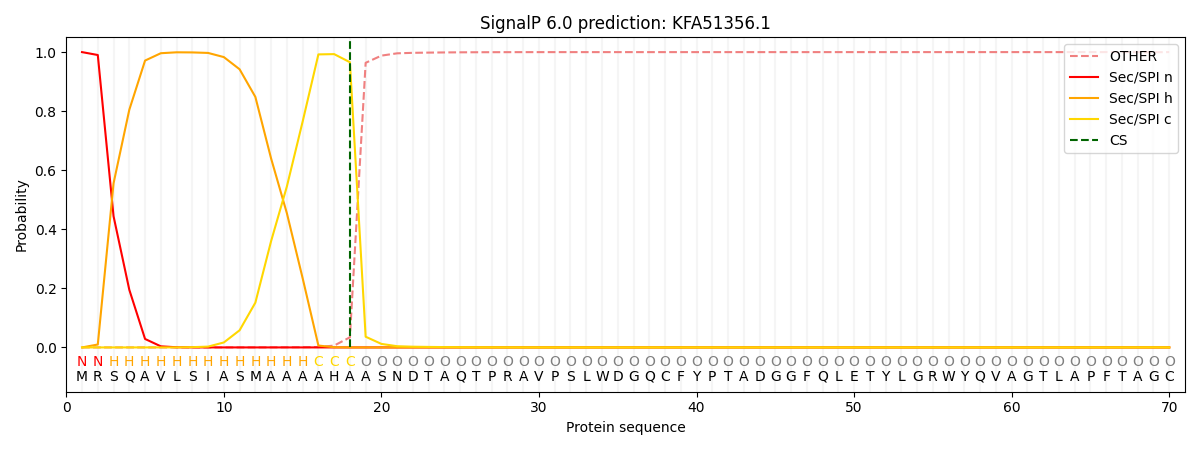
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000639 | 0.999329 | CS pos: 18-19. Pr: 0.9657 |
