You are browsing environment: FUNGIDB
CAZyme Information: KFA47671.1
You are here: Home > Sequence: KFA47671.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Stachybotrys chartarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Stachybotryaceae; Stachybotrys; Stachybotrys chartarum | |||||||||||
| CAZyme ID | KFA47671.1 | |||||||||||
| CAZy Family | AA8 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 131 | 311 | 1.3e-92 | 0.989010989010989 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 214765 | Amb_all | 1.45e-59 | 128 | 313 | 2 | 190 | Amb_all domain. |
| 226384 | PelB | 1.95e-59 | 125 | 375 | 78 | 344 | Pectate lyase [Carbohydrate transport and metabolism]. |
| 366158 | Pec_lyase_C | 9.18e-38 | 140 | 309 | 32 | 211 | Pectate lyase. This enzyme forms a right handed beta helix structure. Pectate lyase is an enzyme involved in the maceration and soft rotting of plant tissue. |
| 395595 | CBM_1 | 1.30e-14 | 20 | 48 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 3.52e-14 | 19 | 52 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.20e-130 | 82 | 376 | 147 | 440 | |
| 2.93e-126 | 81 | 376 | 33 | 327 | |
| 6.56e-124 | 82 | 376 | 112 | 405 | |
| 1.27e-123 | 82 | 376 | 111 | 404 | |
| 1.93e-123 | 85 | 376 | 115 | 406 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.23e-44 | 136 | 376 | 73 | 326 | Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 [Bacillus sp. N16-5],3VMW_A Crystal structure of pectate lyase Bsp165PelA from Bacillus sp. N165 in complex with trigalacturonate [Bacillus sp. N16-5] |
|
| 2.12e-31 | 98 | 286 | 17 | 246 | Chain A, PECTATE LYASE E [Dickeya chrysanthemi] |
|
| 7.27e-30 | 140 | 287 | 130 | 302 | Structure of the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
|
| 4.25e-26 | 138 | 292 | 123 | 301 | Structural insights into the loss of catalytic competence in pectate lyase at low pH [Bacillus subtilis],5X2I_A Polygalacturonate Lyase by Fusing with a Self-assembling Amphipathic Peptide [Bacillus subtilis subsp. subtilis str. 168] |
|
| 5.41e-26 | 138 | 292 | 144 | 322 | Bacillus Subtilis Pectate Lyase [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.44e-61 | 125 | 371 | 74 | 316 | Probable pectate lyase A OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyA PE=3 SV=1 |
|
| 1.44e-61 | 125 | 371 | 74 | 316 | Probable pectate lyase A OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyA PE=3 SV=1 |
|
| 4.42e-60 | 125 | 371 | 74 | 316 | Probable pectate lyase A OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyA PE=3 SV=1 |
|
| 6.97e-60 | 98 | 328 | 45 | 276 | Probable pectate lyase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyB PE=3 SV=1 |
|
| 3.55e-58 | 93 | 371 | 47 | 326 | Pectate lyase B OS=Colletotrichum gloeosporioides OX=474922 GN=PLB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
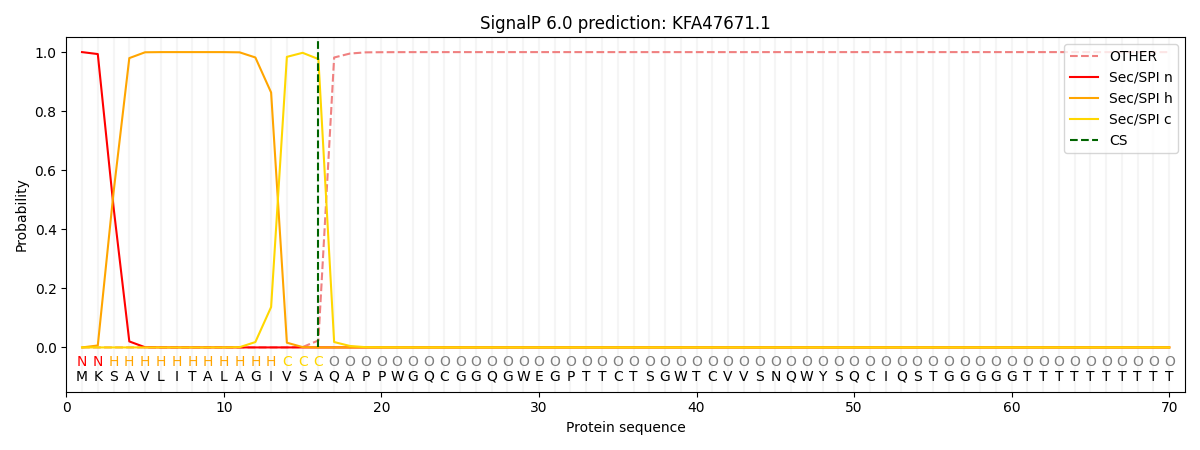
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000309 | 0.999676 | CS pos: 16-17. Pr: 0.9764 |
