You are browsing environment: FUNGIDB
CAZyme Information: KFA45930.1
You are here: Home > Sequence: KFA45930.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
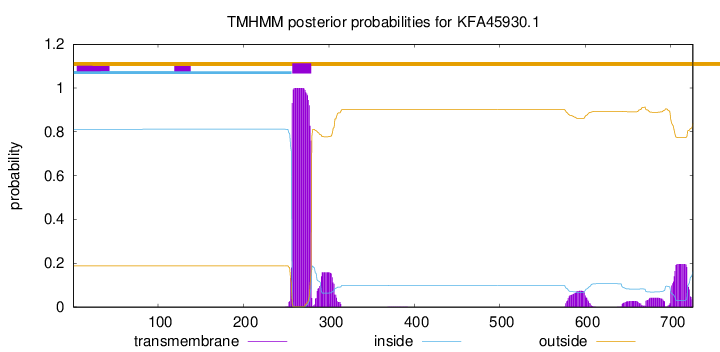
TMHMM annotations
Basic Information help
| Species | Stachybotrys chartarum | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Sordariomycetes; ; Stachybotryaceae; Stachybotrys; Stachybotrys chartarum | |||||||||||
| CAZyme ID | KFA45930.1 | |||||||||||
| CAZy Family | AA3 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT34 | 289 | 553 | 1.1e-80 | 0.9878048780487805 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 368535 | Glyco_transf_34 | 6.61e-77 | 289 | 551 | 3 | 238 | galactosyl transferase GMA12/MNN10 family. This family contains a number of glycosyltransferase enzymes that contain a DXD motif. This family includes a number of C. elegans homologs where the DXD is replaced by DXH. Some members of this family are included in glycosyltransferase family 34. |
| 226211 | COG3686 | 1.76e-06 | 602 | 710 | 20 | 112 | Uncharacterized conserved protein, MAPEG superfamily [Function unknown]. |
| 395893 | MAPEG | 1.57e-05 | 636 | 722 | 48 | 126 | MAPEG family. This family is has been called MAPEG (Membrane Associated Proteins in Eicosanoid and Glutathione metabolism). It includes proteins such as Prostaglandin E synthase. This enzyme catalyzes the synthesis of PGE2 from PGH2 (produced by cyclooxygenase from arachidonic acid). Because of structural similarities in the active sites of FLAP, LTC4 synthase and PGE synthase, substrates for each enzyme can compete with one another and modulate synthetic activity. |
| 227709 | ROM1 | 1.11e-04 | 43 | 227 | 74 | 267 | RhoGEF, Guanine nucleotide exchange factor for Rho/Rac/Cdc42-like GTPases [Signal transduction mechanisms]. |
| 401034 | Rb_C | 0.001 | 118 | 168 | 6 | 53 | Rb C-terminal domain. The Rb C-terminal domain is required for high-affinity binding to E2F-DP complexes and for maximal repression of E2F-responsive promoters, thereby acting as a growth suppressor by blocking the G1-S transition of the cell cycle. This domain has a strand-loop-helix structure, which directly interacts with both E2F1 and DP1, followed by a tail segment that lacks regular secondary structure. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.07e-246 | 135 | 590 | 1 | 455 | |
| 7.24e-245 | 84 | 590 | 29 | 532 | |
| 3.16e-241 | 135 | 590 | 1 | 446 | |
| 1.57e-237 | 135 | 590 | 1 | 459 | |
| 1.57e-237 | 135 | 590 | 1 | 459 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.49e-103 | 295 | 573 | 126 | 390 | Probable alpha-1,6-mannosyltransferase MNN10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=MNN10 PE=1 SV=1 |
|
| 7.72e-86 | 298 | 559 | 82 | 331 | Uncharacterized alpha-1,2-galactosyltransferase C637.06 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC637.06 PE=2 SV=2 |
|
| 3.03e-26 | 316 | 545 | 125 | 333 | Uncharacterized alpha-1,2-galactosyltransferase C1289.13c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1289.13c PE=3 SV=1 |
|
| 9.46e-23 | 319 | 551 | 128 | 339 | Alpha-1,2-galactosyltransferase OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gma12 PE=1 SV=1 |
|
| 1.69e-18 | 319 | 531 | 112 | 311 | Alpha-1,2-galactosyltransferase gmh3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=gmh3 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000055 | 0.000000 |