You are browsing environment: FUNGIDB
CAZyme Information: KEF61504.1
Basic Information
help
| Species |
Exophiala aquamarina
|
| Lineage |
Ascomycota; Eurotiomycetes; ; Herpotrichiellaceae; Exophiala; Exophiala aquamarina
|
| CAZyme ID |
KEF61504.1
|
| CAZy Family |
GT2 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 308 |
AMGV01000002|CGC3 |
32723.03 |
4.2447 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_EaquamarinaCBS119918 |
13118 |
1182545 |
0 |
13118
|
|
| Gene Location |
| Family |
Start |
End |
Evalue |
family coverage |
| GH12 |
169 |
297 |
1.3e-20 |
0.8141025641025641 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 396303
|
Glyco_hydro_12 |
5.66e-15 |
65 |
304 |
3 |
207 |
Glycosyl hydrolase family 12. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 4.72e-95 |
1 |
305 |
1 |
322 |
| 6.27e-95 |
1 |
305 |
1 |
320 |
| 6.27e-95 |
1 |
305 |
1 |
320 |
| 5.61e-93 |
1 |
305 |
1 |
322 |
| 1.59e-92 |
1 |
305 |
1 |
322 |
KEF61504.1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1.81e-13 |
59 |
302 |
30 |
239 |
Xyloglucan-specific endo-beta-1,4-glucanase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=xgeA PE=1 SV=1 |
| 2.70e-11 |
59 |
291 |
31 |
227 |
Xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus niger OX=5061 GN=xgeA PE=1 SV=1 |
| 2.70e-11 |
59 |
291 |
31 |
227 |
Probable xyloglucan-specific endo-beta-1,4-glucanase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=xgeA PE=3 SV=1 |
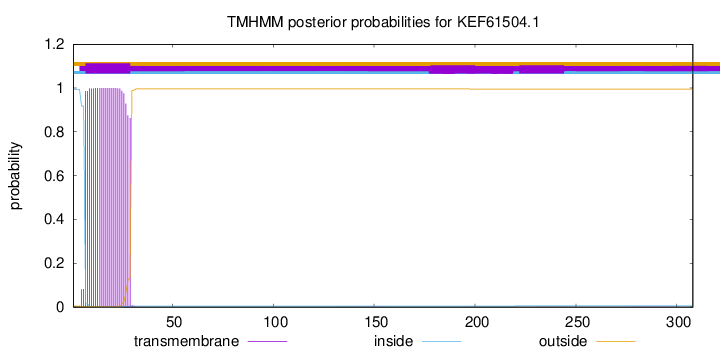
This protein is predicted as OTHER
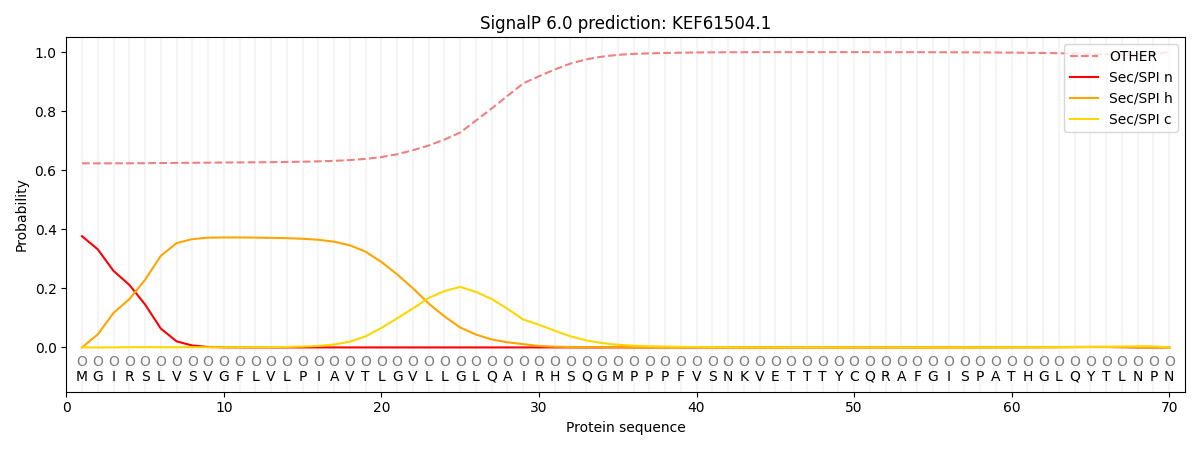
| Other |
SP_Sec_SPI |
CS Position |
| 0.641320 |
0.358677 |
|