You are browsing environment: FUNGIDB
CAZyme Information: KDQ23481.1
You are here: Home > Sequence: KDQ23481.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pleurotus ostreatus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Pleurotaceae; Pleurotus; Pleurotus ostreatus | |||||||||||
| CAZyme ID | KDQ23481.1 | |||||||||||
| CAZy Family | AA5 | |||||||||||
| CAZyme Description | carbohydrate esterase family 15 protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.-:10 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 92 | 413 | 1e-93 | 0.9888475836431226 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 223021 | PHA03247 | 1.49e-07 | 27 | 109 | 2724 | 2815 | large tegument protein UL36; Provisional |
| 223021 | PHA03247 | 2.34e-07 | 36 | 184 | 2702 | 2858 | large tegument protein UL36; Provisional |
| 411345 | gliding_GltJ | 3.27e-05 | 21 | 83 | 396 | 457 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| 237865 | PRK14951 | 3.55e-05 | 22 | 117 | 383 | 492 | DNA polymerase III subunits gamma and tau; Provisional |
| 237030 | kgd | 4.57e-05 | 21 | 104 | 44 | 125 | multifunctional oxoglutarate decarboxylase/oxoglutarate dehydrogenase thiamine pyrophosphate-binding subunit/dihydrolipoyllysine-residue succinyltransferase subunit. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.15e-169 | 88 | 445 | 109 | 466 | |
| 1.95e-166 | 92 | 445 | 116 | 469 | |
| 3.20e-157 | 88 | 445 | 104 | 457 | |
| 4.72e-155 | 92 | 445 | 123 | 481 | |
| 6.69e-155 | 92 | 445 | 123 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.74e-153 | 88 | 445 | 22 | 375 | Chain A, Cip2 [Trichoderma reesei],3PIC_B Chain B, Cip2 [Trichoderma reesei],3PIC_C Chain C, Cip2 [Trichoderma reesei] |
|
| 2.58e-136 | 86 | 445 | 12 | 380 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor],6RU2_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor] |
|
| 6.86e-136 | 86 | 445 | 18 | 386 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant [Cerrena unicolor],6RTV_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant [Cerrena unicolor],6RU1_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X [Cerrena unicolor],6RU1_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X [Cerrena unicolor],6RV7_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR [Cerrena unicolor],6RV7_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR [Cerrena unicolor],6RV9_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR [Cerrena unicolor],6RV9_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR [Cerrena unicolor] |
|
| 3.38e-135 | 86 | 445 | 90 | 458 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor],6RV8_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor] |
|
| 4.21e-132 | 88 | 445 | 55 | 411 | Crystal structure of recombinant glucuronoyl esterase from Sporotrichum thermophile determined at 1.55 A resolution [Thermothelomyces thermophilus ATCC 42464] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.19e-155 | 92 | 445 | 123 | 481 | 4-O-methyl-glucuronoyl methylesterase OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=ge1 PE=1 SV=1 |
|
| 8.24e-151 | 88 | 445 | 107 | 460 | 4-O-methyl-glucuronoyl methylesterase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cip2 PE=1 SV=1 |
|
| 5.33e-145 | 89 | 444 | 140 | 495 | 4-O-methyl-glucuronoyl methylesterase 1 OS=Sodiomyces alcalophilus OX=398408 PE=1 SV=1 |
|
| 1.38e-134 | 86 | 445 | 106 | 474 | 4-O-methyl-glucuronoyl methylesterase OS=Cerrena unicolor OX=90312 PE=1 SV=1 |
|
| 1.47e-132 | 92 | 445 | 43 | 394 | 4-O-methyl-glucuronoyl methylesterase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=Cip2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
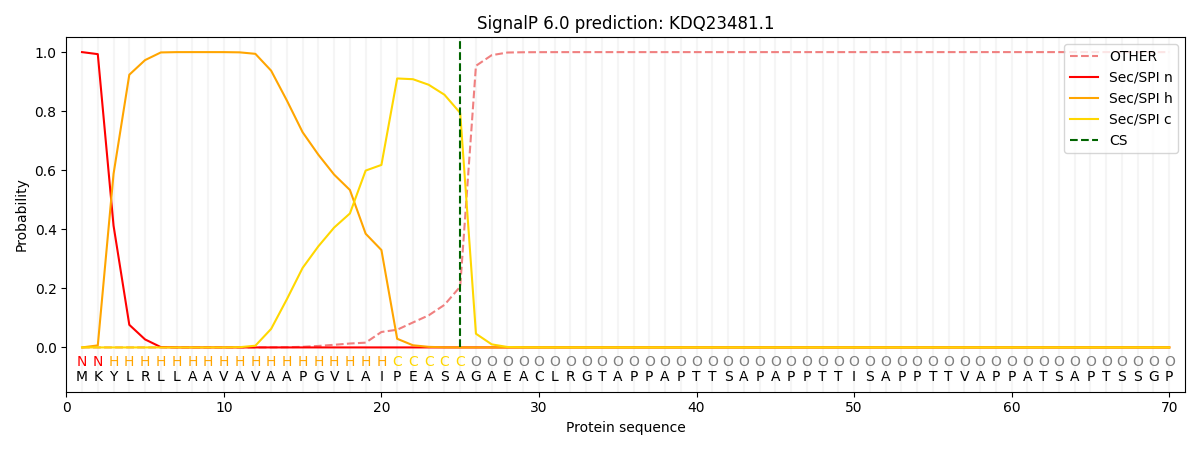
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000315 | 0.999659 | CS pos: 25-26. Pr: 0.7940 |
