You are browsing environment: FUNGIDB
CAZyme Information: KAG6615605.1
Basic Information
help
| Species |
Phytophthora cinnamomi
|
| Lineage |
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi
|
| CAZyme ID |
KAG6615605.1
|
| CAZy Family |
GT1 |
| CAZyme Description |
beta-glucosidase btgE
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PcinnamomiGKB4 |
19981 |
N/A |
0 |
19981
|
|
| Gene Location |
No EC number prediction in KAG6615605.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH17 |
81 |
266 |
1.1e-17 |
0.7106109324758842 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 227625
|
Scw11 |
8.07e-12 |
5 |
255 |
24 |
300 |
Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033
|
Glyco_hydro_17 |
0.001 |
172 |
266 |
220 |
307 |
Glycosyl hydrolases family 17. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 5.12e-57 |
28 |
263 |
23 |
274 |
| 1.03e-22 |
20 |
264 |
15 |
272 |
| 9.51e-20 |
28 |
264 |
21 |
270 |
| 1.29e-18 |
28 |
266 |
19 |
228 |
| 4.26e-18 |
28 |
263 |
94 |
374 |
KAG6615605.1 has no PDB hit.
KAG6615605.1 has no Swissprot hit.
This protein is predicted as SP
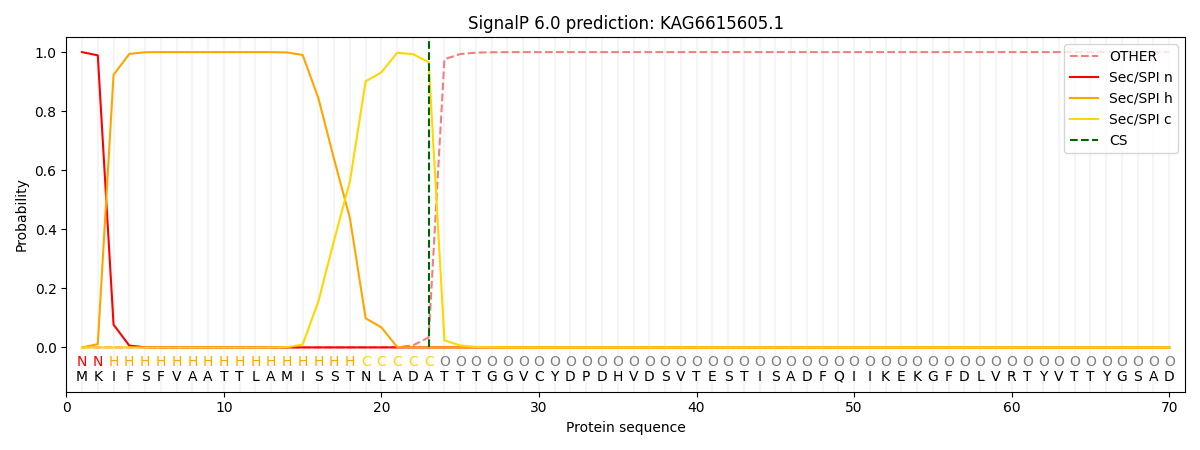
| Other |
SP_Sec_SPI |
CS Position |
| 0.000204 |
0.999782 |
CS pos: 23-24. Pr: 0.9660 |
There is no transmembrane helices in KAG6615605.1.

