You are browsing environment: FUNGIDB
CAZyme Information: KAG6615389.1
You are here: Home > Sequence: KAG6615389.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | KAG6615389.1 | |||||||||||
| CAZy Family | GT1 | |||||||||||
| CAZyme Description | putative polysaccharide lyase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL3 | 29 | 224 | 2.2e-78 | 0.9896907216494846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397360 | Pectate_lyase | 7.62e-86 | 23 | 228 | 2 | 199 | Pectate lyase. |
| 273167 | rad23 | 0.006 | 299 | 362 | 71 | 136 | UV excision repair protein Rad23. All proteins in this family for which functions are known are components of a multiprotein complex used for targeting nucleotide excision repair to specific parts of the genome. In humans, Rad23 complexes with the XPC protein. This family is based on the phylogenomic analysis of JA Eisen (1999, Ph.D. Thesis, Stanford University). [DNA metabolism, DNA replication, recombination, and repair] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.68e-138 | 4 | 252 | 5 | 253 | |
| 5.01e-114 | 5 | 249 | 12 | 258 | |
| 1.16e-112 | 17 | 249 | 28 | 258 | |
| 2.05e-91 | 13 | 260 | 16 | 284 | |
| 2.05e-91 | 13 | 260 | 16 | 284 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.46e-24 | 36 | 168 | 7 | 135 | Crystal Structure Of Pectate Lyase From Bacillus Sp. Strain Ksm-P15. [Bacillus sp. KSM-P15] |
|
| 6.71e-23 | 52 | 249 | 8 | 223 | Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B90_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi] |
|
| 5.17e-22 | 52 | 249 | 126 | 341 | Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B4N_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi],3B8Y_A Chain A, Endo-pectate lyase [Dickeya chrysanthemi],3B8Y_B Chain B, Endo-pectate lyase [Dickeya chrysanthemi] |
|
| 5.50e-20 | 30 | 168 | 6 | 138 | The liganded structure of C. bescii family 3 pectate lyase [Caldicellulosiruptor bescii DSM 6725],4EW9_B The liganded structure of C. bescii family 3 pectate lyase [Caldicellulosiruptor bescii DSM 6725] |
|
| 5.63e-20 | 30 | 168 | 7 | 139 | The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii],3T9G_B The crystal structure of family 3 pectate lyase from Caldicellulosiruptor bescii [Caldicellulosiruptor bescii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.19e-55 | 14 | 245 | 11 | 236 | Probable pectate lyase D OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyD PE=3 SV=1 |
|
| 2.19e-55 | 14 | 245 | 11 | 236 | Probable pectate lyase D OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=plyD PE=3 SV=1 |
|
| 8.78e-55 | 26 | 245 | 37 | 248 | Pectate lyase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyE PE=1 SV=1 |
|
| 1.67e-54 | 11 | 245 | 10 | 235 | Probable pectate lyase D OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyD PE=3 SV=1 |
|
| 1.74e-54 | 26 | 244 | 37 | 247 | Probable pectate lyase E OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
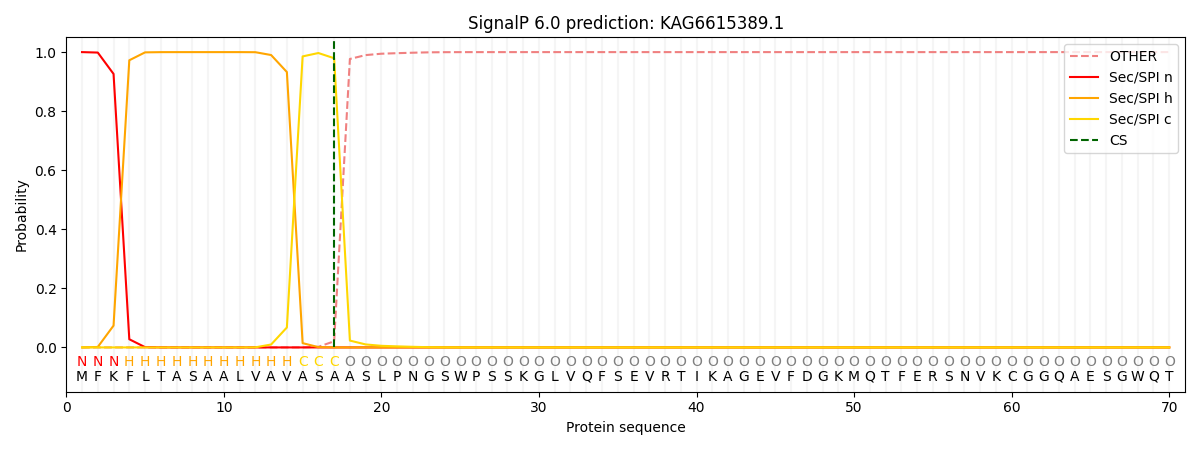
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000206 | 0.999766 | CS pos: 17-18. Pr: 0.9792 |
