You are browsing environment: FUNGIDB
CAZyme Information: KAG6615140.1
Basic Information
help
| Species |
Phytophthora cinnamomi
|
| Lineage |
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi
|
| CAZyme ID |
KAG6615140.1
|
| CAZy Family |
GH81 |
| CAZyme Description |
glycoside hydrolase family 17-like protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PcinnamomiGKB4 |
19981 |
N/A |
0 |
19981
|
|
| Gene Location |
No EC number prediction in KAG6615140.1.
| Family |
Start |
End |
Evalue |
family coverage |
| GH17 |
75 |
318 |
3.9e-18 |
0.9067524115755627 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 227625
|
Scw11 |
1.81e-12 |
20 |
314 |
1 |
304 |
Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 366033
|
Glyco_hydro_17 |
2.53e-07 |
54 |
318 |
12 |
304 |
Glycosyl hydrolases family 17. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.38e-71 |
42 |
320 |
28 |
310 |
| 6.94e-65 |
42 |
327 |
27 |
320 |
| 3.13e-62 |
42 |
322 |
97 |
378 |
| 5.39e-59 |
105 |
334 |
16 |
247 |
| 1.37e-58 |
63 |
330 |
45 |
334 |
KAG6615140.1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 4.75e-08 |
70 |
267 |
325 |
520 |
Probable beta-glucosidase btgE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgE PE=3 SV=1 |
This protein is predicted as SP
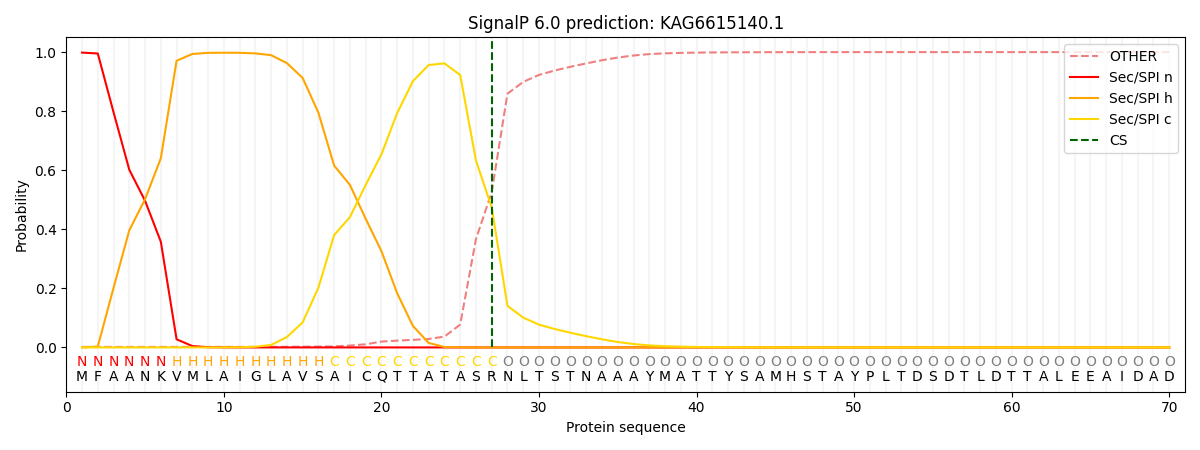
| Other |
SP_Sec_SPI |
CS Position |
| 0.002703 |
0.997263 |
CS pos: 27-28. Pr: 0.4741 |
There is no transmembrane helices in KAG6615140.1.

