You are browsing environment: FUNGIDB
CAZyme Information: KAG6613988.1
You are here: Home > Sequence: KAG6613988.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
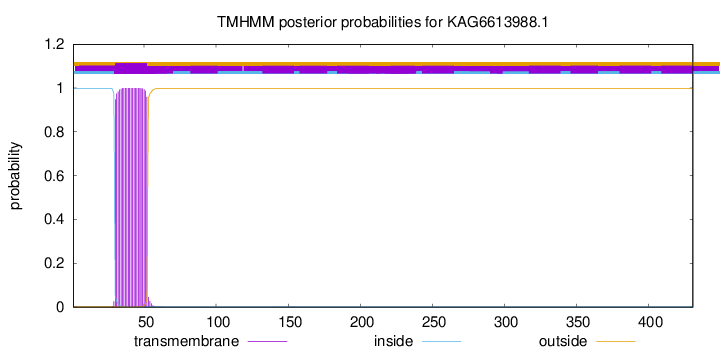
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | KAG6613988.1 | |||||||||||
| CAZy Family | GH7 | |||||||||||
| CAZyme Description | putative choline dehydrogenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 77 | 429 | 5.3e-26 | 0.41585760517799353 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225186 | BetA | 4.99e-15 | 80 | 427 | 28 | 499 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 398739 | GMC_oxred_C | 7.59e-12 | 304 | 428 | 1 | 114 | GMC oxidoreductase. This domain found associated with pfam00732. |
| 235000 | PRK02106 | 5.85e-09 | 77 | 275 | 24 | 249 | choline dehydrogenase; Validated |
| 366272 | GMC_oxred_N | 2.53e-06 | 141 | 280 | 24 | 183 | GMC oxidoreductase. This family of proteins bind FAD as a cofactor. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 9.84e-199 | 1 | 428 | 18 | 606 | |
| 2.01e-09 | 142 | 428 | 83 | 491 | |
| 4.16e-08 | 77 | 315 | 80 | 347 | |
| 6.41e-08 | 334 | 428 | 478 | 573 | |
| 8.51e-08 | 297 | 427 | 460 | 574 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.98e-06 | 77 | 197 | 20 | 139 | Crystal structure of pyridoxine 4-oxidase - pyridoxamine complex [Mesorhizobium loti] |
|
| 9.14e-06 | 77 | 197 | 36 | 155 | Crystal structure of pyridoxine 4-oxidase from Mesorbium loti [Mesorhizobium loti] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999132 | 0.000883 |