You are browsing environment: FUNGIDB
CAZyme Information: KAG6613587.1
You are here: Home > Sequence: KAG6613587.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
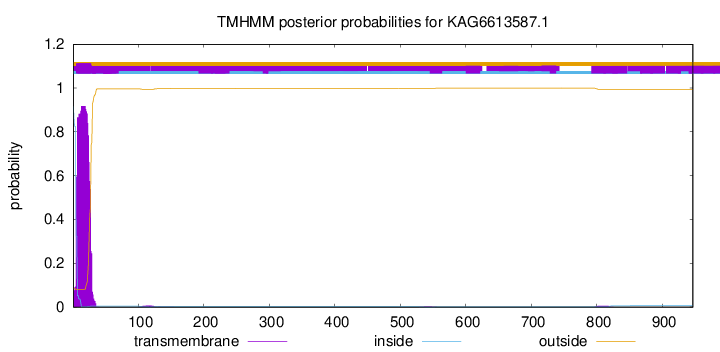
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | KAG6613587.1 | |||||||||||
| CAZy Family | GH7 | |||||||||||
| CAZyme Description | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 72258; End:75704 Strand: + | |||||||||||
Full Sequence Download help
| MWRALGVCAC VCVALAAALS PGFNAPMPYY TYPLLEILHP LNDMLLAQTE LQVEIEVRAE | 60 |
| LLSGGLQRSR VCVLMQPAYV PPDVALDPGA GELDESCFDQ SLNYTTFHVA GLVPGLSYAL | 120 |
| TVGLETNGNM VALSSRTFSV GSILLPGVDG RLSVDDALEA GAQLHNARDR ATASRIYRYV | 180 |
| LEIFPDHPQA QHLLGLALYQ DGETDRALPY LQRAVQANES DENFHNSLGI CLKSLGRVDE | 240 |
| AVRHYRRALE LNPMQVQASI NLGDAMQSQG KWEEALKEYT KVAKIPMHVL ESHTEPGKAE | 300 |
| HLAKDATGRR CELIRVTDGW YHADRCLNDA LERWPDEPVF HNDRGNLLAN AGQFETALDE | 360 |
| YQRSSDLGLL AGKLNLADTL EALGETQKCI DMYDQILGKE ILDRFHPRTR IMVMKATVLP | 420 |
| RVLPSTQKEI DDSRDRFERE VEALLLNLDS LEKTEVDPNR VSLSTGITLT AHNRNNRELK | 480 |
| AAMGYLYWQL LYQRRMVRED YVASYGIVPL PYTQRSEDIN RPAIGPRRLR VGFVSRYIFN | 540 |
| SAVGLYMSEL IPMFDREKYE IIVFAIAQSK SMKVVKEIEA ITETIIALPK DVRIAREEIR | 600 |
| AWHMDVLIYP ELGMDKTTYF VSLDRLAKVQ AVWWGNADTS GVPTMDYYLT SEHEHPNFNS | 660 |
| HYSEAVYQFR GMGIFHKLPD LPKKSINRDQ VRRAIEERFD IPSDFHFYLA IESIIHIHPD | 720 |
| FDAAVAKIFE ADKKAHFFLL STSNRKVWRS QLQERMESAG VDPHRLHFLS DVDQKQEIML | 780 |
| MRAADAVVAS LHMTRPRASL QAFAAGVPVV TFPSELWASR ITYGFYQQMG VNDLVAASLD | 840 |
| EYVALTVKLA TDMAFHKKME QLITRNRSKL SEDVQAVQEW EKFFDFAGRQ IFPSGEQKPG | 900 |
| SNWDWGQTDG LEGIDYWGQI EGVENVVEWS QIEKSDGVID RKLVEKE | 947 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT41 | 313 | 882 | 2.1e-66 | 0.5319148936170213 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226428 | Spy | 6.49e-31 | 511 | 884 | 245 | 611 | Predicted O-linked N-acetylglucosamine transferase, SPINDLY family [Posttranslational modification, protein turnover, chaperones]. |
| 276809 | TPR | 1.55e-20 | 188 | 284 | 1 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 276809 | TPR | 5.00e-16 | 158 | 250 | 5 | 97 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
| 411346 | social_mot_Tgl | 7.44e-14 | 194 | 282 | 37 | 126 | social motility TPR repeat lipoprotein Tgl. Social motility in delta-proteobacterial species such as Myxococcus xanthus depends on a type VI pilus, which in turn depends on assembly of the PilQ secretin complex. Tgl, a tetratricopeptide repeat (TPR) outer membrane lipoprotein, is required for PilQ assembly. |
| 276809 | TPR | 2.32e-12 | 225 | 363 | 4 | 94 | Tetratricopeptide repeat. The Tetratricopeptide repeat (TPR) typically contains 34 amino acids and is found in a variety of organisms including bacteria, cyanobacteria, yeast, fungi, plants, and humans. It is present in a variety of proteins including those involved in chaperone, cell-cycle, transcription, and protein transport complexes. The number of TPR motifs varies among proteins. Those containing 5-6 tandem repeats generate a right-handed helical structure with an amphipathic channel that is thought to accommodate an alpha-helix of a target protein. It has been proposed that TPR proteins preferentially interact with WD-40 repeat proteins, but in many instances several TPR-proteins seem to aggregate to multi-protein complexes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| UIZ20641.1|GT41 | 0.0 | 18 | 914 | 21 | 933 |
| BAZ61948.1|GT41 | 9.34e-61 | 332 | 887 | 365 | 884 |
| BAZ15802.1|GT41 | 9.34e-61 | 332 | 887 | 365 | 884 |
| BBO73735.1|GT41 | 3.27e-58 | 397 | 887 | 360 | 814 |
| BBO66743.1|GT41 | 4.93e-56 | 413 | 885 | 458 | 899 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5DJS_A | 4.18e-16 | 486 | 865 | 121 | 491 | Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_B Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_C Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum],5DJS_D Thermobaculum terrenum O-GlcNAc transferase mutant - K341M [Thermobaculum terrenum] |
| 5NPS_A | 1.04e-07 | 684 | 884 | 493 | 697 | The human O-GlcNAc transferase in complex with a bisubstrate inhibitor [Homo sapiens] |
| 5NPR_A | 1.04e-07 | 684 | 884 | 492 | 696 | The human O-GlcNAc transferase in complex with a thiol-linked bisubstrate inhibitor [Homo sapiens] |
| 5HGV_A | 1.04e-07 | 684 | 884 | 494 | 698 | Structure of an O-GlcNAc transferase point mutant, D554N in complex with peptide [Homo sapiens],5HGV_C Structure of an O-GlcNAc transferase point mutant, D554N in complex with peptide [Homo sapiens] |
| 6IBO_A | 1.04e-07 | 684 | 884 | 498 | 702 | Catalytic deficiency of O-GlcNAc transferase leads to X-linked intellectual disability [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|P56558|OGT1_RAT | 2.70e-07 | 684 | 884 | 806 | 1010 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Rattus norvegicus OX=10116 GN=Ogt PE=1 SV=1 |
| sp|Q8CGY8|OGT1_MOUSE | 2.71e-07 | 684 | 884 | 816 | 1020 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Mus musculus OX=10090 GN=Ogt PE=1 SV=2 |
| sp|Q9M8Y0|SEC_ARATH | 6.00e-07 | 525 | 850 | 588 | 912 | Probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC OS=Arabidopsis thaliana OX=3702 GN=SEC PE=1 SV=1 |
| sp|O15294|OGT1_HUMAN | 6.12e-07 | 684 | 884 | 816 | 1020 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Homo sapiens OX=9606 GN=OGT PE=1 SV=3 |
| sp|P81436|OGT1_RABIT | 6.12e-07 | 684 | 884 | 816 | 1020 | UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit OS=Oryctolagus cuniculus OX=9986 GN=OGT PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
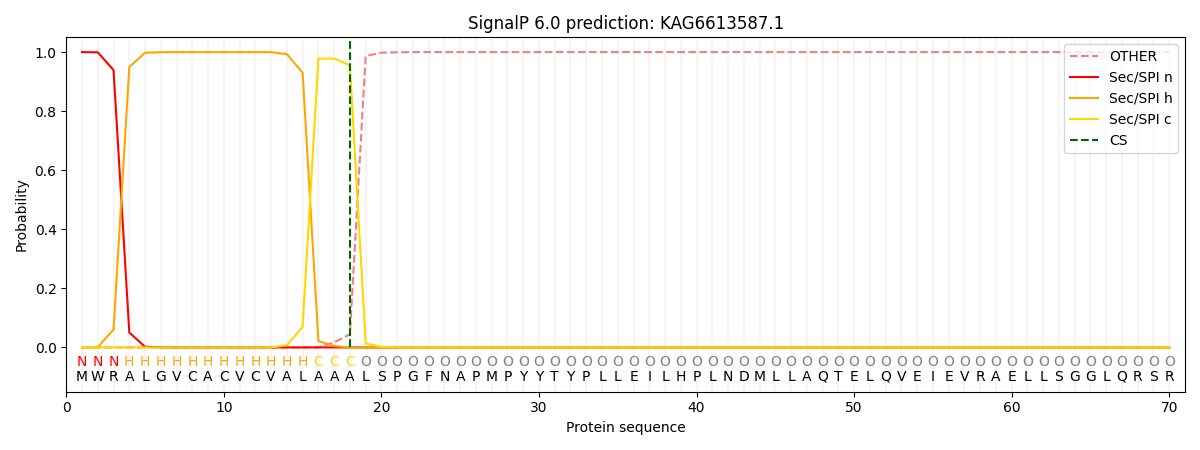
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000232 | 0.999751 | CS pos: 18-19. Pr: 0.9557 |