You are browsing environment: FUNGIDB
CAZyme Information: KAG6612846.1
You are here: Home > Sequence: KAG6612846.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | KAG6612846.1 | |||||||||||
| CAZy Family | GH53 | |||||||||||
| CAZyme Description | histone acetyltransferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.1.99.18:3 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA3 | 20 | 549 | 1.5e-104 | 0.9744525547445255 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 99938 | Bromo_plant1 | 1.22e-42 | 1884 | 1979 | 3 | 98 | Bromodomain, uncharacterized subfamily specific to plants. Might function as a global transcription factor. Bromodomains are 110 amino acid long domains, that are found in many chromatin associated proteins. Bromodomains can interact specifically with acetylated lysine. |
| 225186 | BetA | 4.20e-36 | 22 | 557 | 1 | 539 | Choline dehydrogenase or related flavoprotein [Lipid transport and metabolism, General function prediction only]. |
| 197636 | BROMO | 2.01e-30 | 1882 | 1983 | 8 | 107 | bromo domain. |
| 400497 | HAT_KAT11 | 2.37e-29 | 2229 | 2433 | 6 | 236 | Histone acetylation protein. Histone acetylation is required in many cellular processes including transcription, DNA repair, and chromatin assembly. This family contains the fungal KAT11 protein (previously known as RTT109) which is required for H3K56 acetylation. Loss of KAT11 results in the loss of H3K56 acetylation, both on bulk histone and on chromatin. KAT11 and H3K56 acetylation appear to correlate with actively transcribed genes and associate with the elongating form of Pol II in yeast. This family also incorporates the p300/CBP histone acetyltransferase domain which has different catalytic properties and cofactor regulation to KAT11. |
| 99931 | Bromo_BDF1_2_II | 2.10e-28 | 1887 | 1980 | 6 | 102 | Bromodomain. BDF1/BDF2 like subfamily, restricted to fungi, repeat II. BDF1 and BDF2 are yeast transcription factors involved in the expression of a wide range of genes, including snRNAs; they are required for sporulation and DNA repair and protect histone H4 from deacetylation. Bromodomains are 110 amino acid long domains, that are found in many chromatin associated proteins. Bromodomains can interact specifically with acetylated lysine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.31e-278 | 1 | 588 | 1 | 582 | |
| 1.63e-134 | 13 | 533 | 7 | 526 | |
| 5.31e-133 | 28 | 588 | 23 | 577 | |
| 9.70e-120 | 22 | 588 | 17 | 577 | |
| 1.05e-119 | 8 | 546 | 4 | 544 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.71e-85 | 1902 | 2716 | 39 | 615 | Crystal Structure of the Catalytic Core of CBP [Mus musculus],5U7G_B Crystal Structure of the Catalytic Core of CBP [Mus musculus] |
|
| 4.57e-78 | 1902 | 2438 | 30 | 478 | Cryo-EM structure of p300-p53 protein complex [Homo sapiens],6K4N_A Cryo-EM structure of p300 [Homo sapiens] |
|
| 7.17e-78 | 1902 | 2438 | 31 | 479 | Transcription factor dimerization activates the p300 acetyltransferase [Homo sapiens],6GYR_B Transcription factor dimerization activates the p300 acetyltransferase [Homo sapiens],6GYR_C Transcription factor dimerization activates the p300 acetyltransferase [Homo sapiens],6GYR_D Transcription factor dimerization activates the p300 acetyltransferase [Homo sapiens] |
|
| 1.16e-76 | 1902 | 2433 | 43 | 486 | Structural basis for autoinhibition of the acetyltransferase activity of p300 [Homo sapiens],4BHW_B Structural basis for autoinhibition of the acetyltransferase activity of p300 [Homo sapiens],5LKT_A Crystal structure of the p300 acetyltransferase catalytic core with butyryl-coenzyme A. [Homo sapiens],5LKU_A Crystal structure of the p300 acetyltransferase catalytic core with coenzyme A. [Homo sapiens],5LKX_A Crystal structure of the p300 acetyltransferase catalytic core with propionyl-coenzyme A. [Homo sapiens],5LKZ_A Crystal structure of the p300 acetyltransferase catalytic core with crotonyl-coenzyme A. [Homo sapiens] |
|
| 6.19e-60 | 2004 | 2433 | 8 | 362 | Chain A, Histone acetyltransferase p300 [Homo sapiens],7VHY_B Chain B, Histone acetyltransferase p300 [Homo sapiens],7VHZ_A Chain A, Histone acetyltransferase p300 [Homo sapiens],7VHZ_B Chain B, Histone acetyltransferase p300 [Homo sapiens],7VI0_A Chain A, Histone acetyltransferase p300 [Homo sapiens],7VI0_B Chain B, Histone acetyltransferase p300 [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 9.39e-176 | 1998 | 2849 | 955 | 1696 | Histone acetyltransferase HAC12 OS=Arabidopsis thaliana OX=3702 GN=HAC12 PE=3 SV=2 |
|
| 2.33e-164 | 1998 | 2849 | 946 | 1687 | Histone acetyltransferase HAC1 OS=Arabidopsis thaliana OX=3702 GN=HAC1 PE=1 SV=2 |
|
| 8.36e-156 | 1998 | 2849 | 959 | 1659 | Probable histone acetyltransferase HAC-like 1 OS=Oryza sativa subsp. japonica OX=39947 GN=Os02g0137500 PE=3 SV=2 |
|
| 6.07e-152 | 1998 | 2849 | 927 | 1659 | Histone acetyltransferase HAC5 OS=Arabidopsis thaliana OX=3702 GN=HAC5 PE=2 SV=1 |
|
| 3.91e-147 | 1999 | 2849 | 722 | 1461 | Histone acetyltransferase HAC4 OS=Arabidopsis thaliana OX=3702 GN=HAC4 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
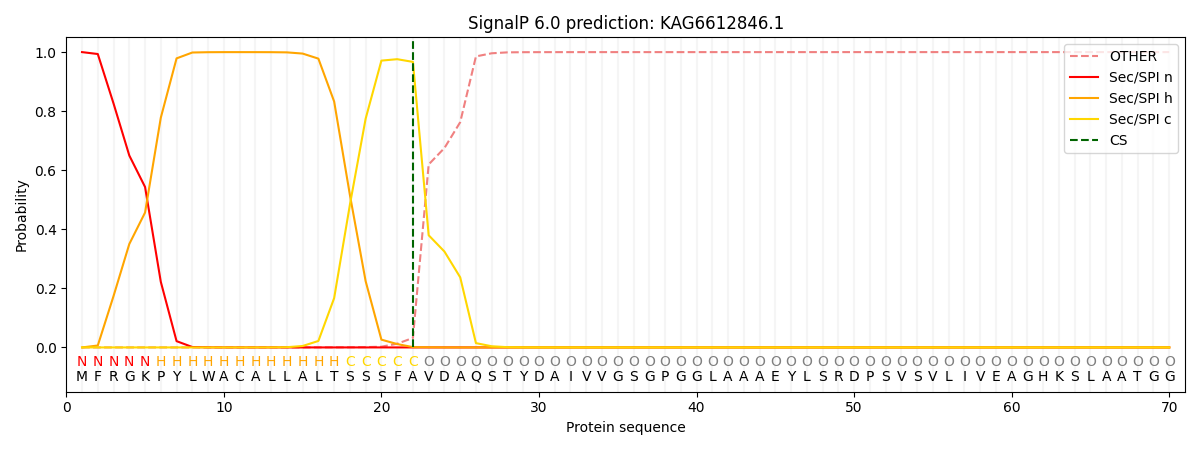
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000596 | 0.999376 | CS pos: 22-23. Pr: 0.9668 |
