You are browsing environment: FUNGIDB
CAZyme Information: KAG6612293.1
You are here: Home > Sequence: KAG6612293.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | KAG6612293.1 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 1.14.99.-:4 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA17 | 14 | 274 | 2.7e-60 | 0.9673469387755103 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 183854 | PRK13042 | 4.58e-05 | 212 | 284 | 17 | 88 | superantigen-like protein SSL4; Reviewed. |
| 411470 | opacity_OapA | 0.007 | 198 | 288 | 169 | 266 | opacity-associated protein OapA. This family consists of full-length homologs to OapA, opacity-associated protein A as described in Haemophilus influenzae. OapA shares a C-terminal homology domain, called the OapA domain, with the Escherichia coli protein YtfB, which is now known to bind peptidoglycan through its OapA domain and to act as a cell division protein. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.56e-72 | 1 | 211 | 1 | 203 | |
| 1.56e-72 | 1 | 211 | 1 | 203 | |
| 3.70e-72 | 1 | 211 | 1 | 204 | |
| 4.43e-72 | 1 | 211 | 1 | 203 | |
| 4.43e-72 | 1 | 211 | 1 | 203 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.97e-61 | 26 | 204 | 1 | 172 | Chain A, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4],6Z5Y_B Chain B, Lytic Polysaccharide Monooxygenase [Phytophthora infestans T30-4] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
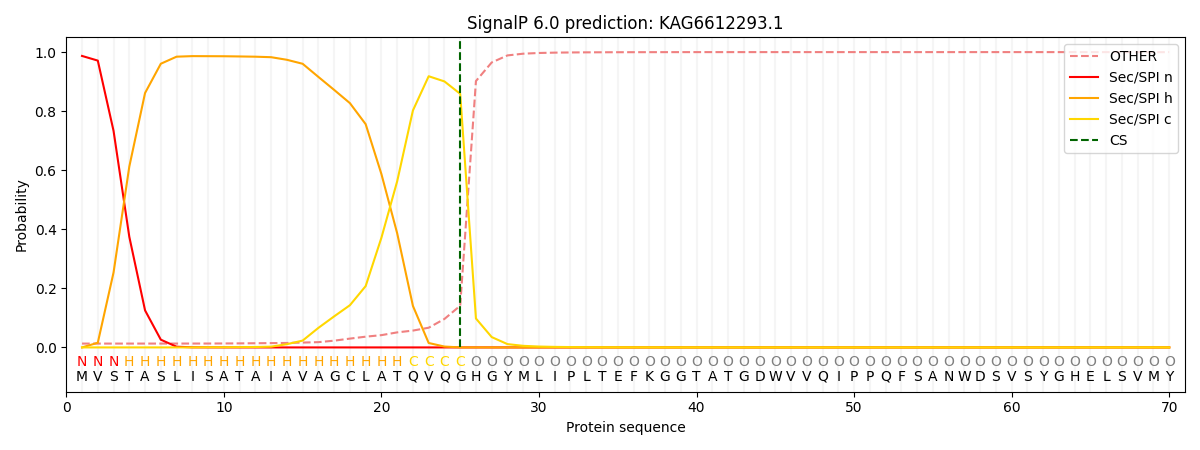
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.016809 | 0.983185 | CS pos: 25-26. Pr: 0.8587 |
