You are browsing environment: FUNGIDB
KAG6609554.1
Basic Information
help
Species
Phytophthora cinnamomi
Lineage
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi
CAZyme ID
KAG6609554.1
CAZy Family
GH3
CAZyme Description
RxLR-like protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
458
49578.52
4.2593
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_PcinnamomiGKB4
19981
N/A
0
19981
Gene Location
Family
Start
End
Evalue
family coverage
GH30
14
445
1.1e-88
0.9013157894736842
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
405300
Glyco_hydr_30_2
1.71e-10
70
157
97
182
O-Glycosyl hydrolase family 30.
more
399741
Alpha-L-AF_C
0.008
313
441
63
192
Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
1.55e-135
3
447
73
496
1.15e-103
3
449
79
496
1.48e-101
3
450
80
498
3.66e-99
3
450
80
498
3.66e-99
3
450
80
498
KAG6609554.1 has no PDB hit.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
1.54e-73
13
452
72
478
Endo-beta-1,6-galactanase OS=Hypocrea rufa OX=5547 GN=6GAL PE=1 SV=1
more
5.82e-06
16
166
106
271
Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1
more
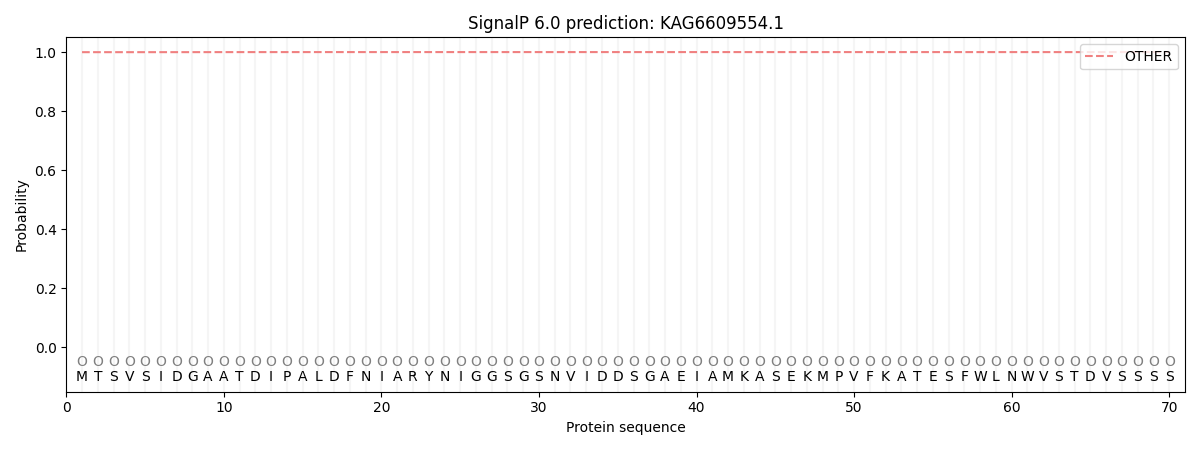
This protein is predicted as OTHER
Other
SP_Sec_SPI
CS Position
0.999841
0.000205
There is no transmembrane helices in KAG6609554.1.