You are browsing environment: FUNGIDB
CAZyme Information: KAG6608882.1
Basic Information
help
| Species |
Phytophthora cinnamomi
|
| Lineage |
Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi
|
| CAZyme ID |
KAG6608882.1
|
| CAZy Family |
GH28 |
| CAZyme Description |
Major Facilitator Superfamily (MFS)
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 165 |
|
17923.60 |
6.0633 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_PcinnamomiGKB4 |
19981 |
N/A |
0 |
19981
|
|
| Gene Location |
No EC number prediction in KAG6608882.1.
| Family |
Start |
End |
Evalue |
family coverage |
| AA17 |
1 |
144 |
7.7e-47 |
0.5755102040816327 |
KAG6608882.1 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.67e-64 |
1 |
165 |
107 |
276 |
| 1.10e-57 |
1 |
165 |
140 |
327 |
| 2.04e-56 |
1 |
165 |
70 |
258 |
| 5.87e-56 |
1 |
165 |
107 |
295 |
| 6.75e-54 |
1 |
96 |
107 |
205 |
KAG6608882.1 has no PDB hit.
KAG6608882.1 has no Swissprot hit.
This protein is predicted as OTHER
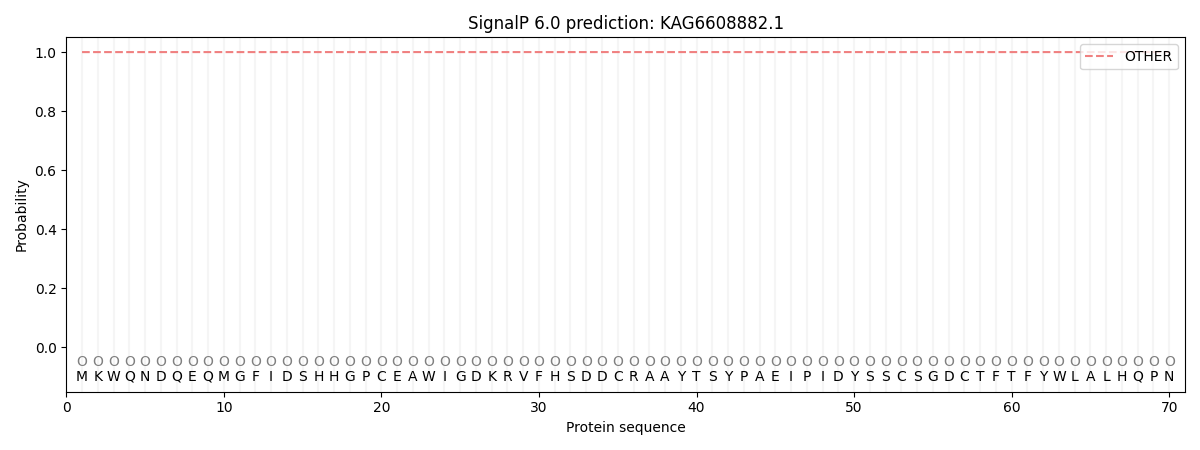
| Other |
SP_Sec_SPI |
CS Position |
| 1.000054 |
0.000002 |
|
There is no transmembrane helices in KAG6608882.1.

