You are browsing environment: FUNGIDB
CAZyme Information: KAG6574623.1
You are here: Home > Sequence: KAG6574623.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | KAG6574623.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | Alpha-N-acetylglucosaminidase (NAGLU) | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 503381; End:505869 Strand: + | |||||||||||
Full Sequence Download help
| MLSLATLLVL LGARSEASID LLKRDAAAAD VVAATRGLIH RRLGARFNDQ IALRVLPSDA | 60 |
| DGLDVFELGS AGDKLEVAAN SATAMAYGLH WYLKTALHTQ TDWDDHKLQL PYVLPKLDKP | 120 |
| VRHKRSAKFS YYQNVCTVSY SSWAWGWQKW EKHIDWMALN GINMPLAFTG QEKVWQDTFQ | 180 |
| KHYNVSSAGL NKFFAGSAFL AWGRMGNLRG SWVEGPLPQA FIDGQHELQL KILSRMREFG | 240 |
| MVPALPAFAG HVPEELKTLY PNAKFTRSPN WGGFSDEFCC VYMLDPQDPL YHEIGKTFLE | 300 |
| EQRALYDYTS SLYQCDTYNE MDPDFTDPAE LQAASRAVIN SMTAADPNAV WLIQGWLFRS | 360 |
| AACVEQDGQL LRQKLDLLPN RASDGTMVGV GLTMEGIFQN YVVYDLTLQM AWVDAPLDMD | 420 |
| EWVPSFAAQR YHSQDAHTER AWGFLLQSVY NRTLGYGGVT KNLVCLIPHW KLVRDGFMPT | 480 |
| LITYDPMDIA RAWKELLLAG TELHAVDTYR HDLVDVTRQF LSDHFMAQYL HLKDLYTGKE | 540 |
| VAADQLCAWT DRMLVTIERL DEILATNDDF LLGNWIADAR ALADGFETTE GSNLQDYYEY | 600 |
| EARNQVTRWG DNNSESIHDY AGKEWAGLVN GYYLPRWRMW LTEVCQSYTE KREIDDAALK | 660 |
| KARIDFELGW QLSHEPYPTT ATGDTLAVSK RIFDEFAGAN DFQAPWSWRL AGYFAHTQAL | 720 |
| A | 721 |
Enzyme Prediction help
| EC | 3.2.1.50:13 | 3.2.1.50:13 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH89 | 72 | 361 | 2.6e-108 | 0.4238310708898944 |
| GH89 | 379 | 693 | 1.6e-81 | 0.45852187028657615 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 398659 | NAGLU | 7.37e-136 | 131 | 412 | 2 | 331 | Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This central domain has a tim barrel fold. |
| 404009 | NAGLU_C | 3.52e-95 | 422 | 690 | 1 | 258 | Alpha-N-acetylglucosaminidase (NAGLU) C-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This C-terminal domain has an all alpha helical fold. |
| 404008 | NAGLU_N | 1.29e-22 | 36 | 115 | 4 | 81 | Alpha-N-acetylglucosaminidase (NAGLU) N-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This N-terminal domain has an alpha-beta fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIG56008.1|GH89 | 7.28e-273 | 30 | 697 | 26 | 739 |
| AIG55486.1|GH89 | 2.22e-271 | 30 | 705 | 23 | 738 |
| AIG56414.1|GH89 | 2.38e-210 | 30 | 699 | 32 | 751 |
| AIG56322.1|GH89 | 1.14e-192 | 30 | 696 | 31 | 747 |
| AIG55749.1|GH89 | 2.14e-187 | 30 | 696 | 29 | 748 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4XWH_A | 3.18e-131 | 33 | 693 | 7 | 708 | Crystal structure of the human N-acetyl-alpha-glucosaminidase [Homo sapiens] |
| 2VC9_A | 2.57e-69 | 36 | 692 | 175 | 872 | Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with 2-acetamido-1,2-dideoxynojirmycin [Clostridium perfringens],2VCA_A Family 89 glycoside hydrolase from Clostridium perfringens in complex with beta-N-acetyl-D-glucosamine [Clostridium perfringens],2VCB_A Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc [Clostridium perfringens],2VCC_A Family 89 Glycoside Hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 7MFK_A | 2.83e-69 | 36 | 692 | 183 | 880 | Chain A, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124],7MFL_A Chain A, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124] |
| 4A4A_A | 2.25e-68 | 36 | 692 | 198 | 895 | CpGH89 (E483Q, E601Q), from Clostridium perfringens, in complex with its substrate GlcNAc-alpha-1,4-galactose [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q9FNA3|NAGLU_ARATH | 2.17e-138 | 34 | 696 | 48 | 804 | Alpha-N-acetylglucosaminidase OS=Arabidopsis thaliana OX=3702 GN=NAGLU PE=2 SV=1 |
| sp|P54802|ANAG_HUMAN | 3.08e-130 | 33 | 693 | 30 | 731 | Alpha-N-acetylglucosaminidase OS=Homo sapiens OX=9606 GN=NAGLU PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
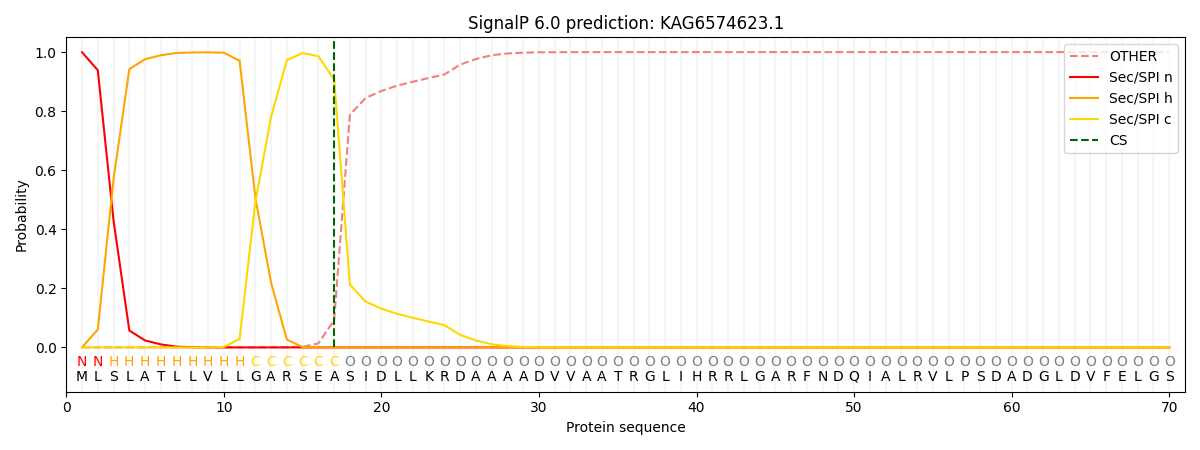
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001041 | 0.998926 | CS pos: 17-18. Pr: 0.9072 |
