You are browsing environment: FUNGIDB
CAZyme Information: KAG6572617.1
You are here: Home > Sequence: KAG6572617.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phytophthora cinnamomi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora cinnamomi | |||||||||||
| CAZyme ID | KAG6572617.1 | |||||||||||
| CAZy Family | AA1 | |||||||||||
| CAZyme Description | mannan endo-1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 511789; End:513206 Strand: + | |||||||||||
Full Sequence Download help
| MKVFTSAVVL GSLAAQVANA GFVKTSGTAF EVDGKPFNVF GTNAYWASEI NWSEANLATI | 60 |
| FKTMATNDLT VCRTMGFADL TTVGTAPYNI VYQLWENGTP TINTKDNGLG YFDKVVAAAK | 120 |
| AAGVKLVVPL VNNWSDYGGM DVYVKQLGGK YHDDFYTNKK VKAAYKKYIA TFVKRYKKED | 180 |
| TIMTWELCNE CRCAGTGGGL PESGSCTTKT INAWMTEMSA YIKSLDSNHL VATGSEGFFN | 240 |
| TDSSVYLYSG LSGVDFDANL AIKSIDYGAY HTYPDGWNVN ANEYVSWGTK WINDHVASGK | 300 |
| KAGKPVVMEE YGVKNHNASV YKAWSDAVYA AGSSMQYWEF GLESLKTYRG DYTIYDTDAI | 360 |
| FKSAIVPAAK KFKTRRSSA | 379 |
Enzyme Prediction help
| EC | 3.2.1.78:7 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 56 | 340 | 4.2e-84 | 0.9896193771626297 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226444 | COG3934 | 3.14e-26 | 126 | 333 | 83 | 270 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| 395098 | Cellulase | 3.27e-05 | 172 | 338 | 113 | 266 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRW08259.1|CBM1|GH5_7 | 6.55e-106 | 18 | 359 | 96 | 434 |
| EPQ57046.1|GH5_7|3.2.1.78 | 2.35e-105 | 1 | 361 | 6 | 365 |
| ABG79370.1|CBM1|GH5_7|3.2.1.78 | 5.02e-105 | 21 | 334 | 102 | 416 |
| AGV52632.1|CBM1|GH5_7 | 2.37e-104 | 18 | 332 | 97 | 408 |
| QRV79289.1|GH5_7 | 2.53e-103 | 19 | 359 | 29 | 369 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4AWE_A | 2.39e-82 | 21 | 371 | 5 | 380 | The Crystal Structure of Chrysonilia sitophila endo-beta-D-1,4- mannanase [Neurospora sitophila] |
| 4QP0_A | 9.27e-81 | 20 | 328 | 2 | 313 | Crystal Structure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| 3PZ9_A | 6.00e-69 | 32 | 356 | 22 | 361 | Native structure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 1QNO_A | 3.13e-68 | 20 | 341 | 2 | 309 | Chain A, ENDO-1,4-B-D-MANNANASE [Trichoderma reesei],1QNP_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNQ_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNR_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei],1QNS_A Chain A, Endo-1,4-b-d-mannanase [Trichoderma reesei] |
| 6TN6_A | 2.29e-66 | 32 | 356 | 8 | 347 | X-ray structure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q5AZ53|MANC_EMENI | 9.88e-84 | 18 | 371 | 18 | 391 | Mannan endo-1,4-beta-mannosidase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manC PE=1 SV=1 |
| sp|Q0C8J3|MANC_ASPTN | 1.91e-82 | 2 | 373 | 7 | 400 | Probable mannan endo-1,4-beta-mannosidase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=manC PE=3 SV=1 |
| sp|Q5AVP1|MAND_EMENI | 3.99e-75 | 14 | 355 | 18 | 348 | Mannan endo-1,4-beta-mannosidase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manD PE=3 SV=1 |
| sp|A1DBV1|MANF_NEOFI | 4.01e-75 | 18 | 366 | 110 | 450 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=manF PE=3 SV=1 |
| sp|B0Y9E7|MANF_ASPFC | 5.98e-73 | 19 | 355 | 94 | 421 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=manF PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
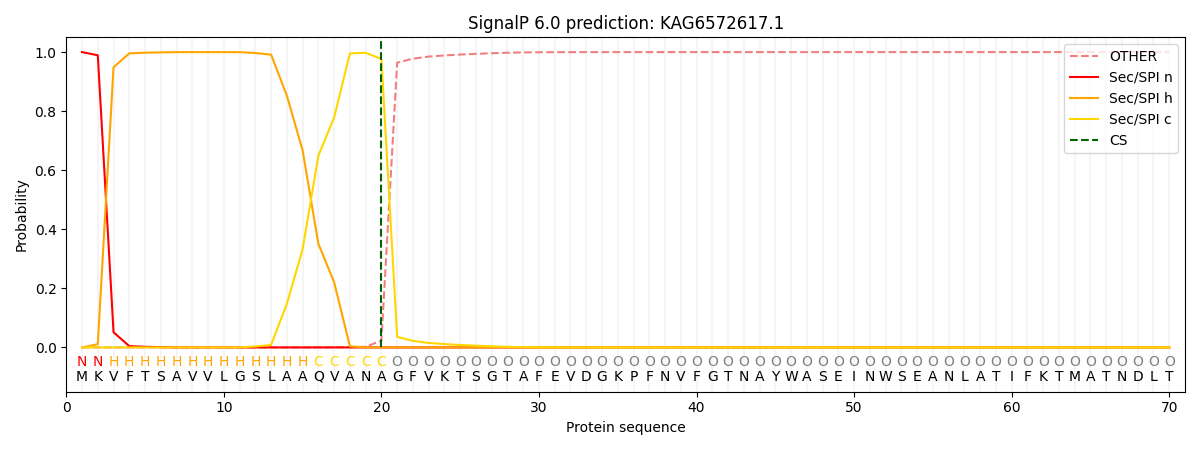
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000221 | 0.999758 | CS pos: 20-21. Pr: 0.9771 |
