You are browsing environment: FUNGIDB
CAZyme Information: KAG5519850.1
You are here: Home > Sequence: KAG5519850.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pneumocystis sp. 'macacae' | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis sp. 'macacae' | |||||||||||
| CAZyme ID | KAG5519850.1 | |||||||||||
| CAZy Family | GT48 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 320056; End:330953 Strand: - | |||||||||||
Full Sequence Download help
| MKQPVKTHKN GKESSKGLLG SSTFNISKQN NSDVSKFKFG AFGSIRERIF SSVMIESLEF | 60 |
| DGLSPDLVVI FKNISKKDTT TKLKALDDIS TWIDDNPGKL NENNFLEIWS YLYPRLTIDA | 120 |
| DRQVRLSCHH VHHKIYNESG KRIAKFVKKI IGSWIASTFD NDRAVKKAAI ESFNTVFKTS | 180 |
| EKRLTVKKVY HKYILNFNKN VILKELPSTL SDLRYVSQEI SEVRYFKIIS SILASLSDII | 240 |
| LNLKLEEIKE EEDSYIEIVS SETFWQFLYQ SDSDVRKYFY ELIKNISNKW PELLMSQKNM | 300 |
| IKKSITKLME NADHLFISVT LSTMEVFYLV CPEIWEHDFF GDYQTFSYFK KLILNNNFNS | 360 |
| EQFWIYFYDL IIKIPLNPHK NNFKIYLNDF IDIIWEGVHK KTKNNIESAF EVFFKIICFY | 420 |
| GKCFKNMDDI FSILEKKVSD IFKLYFNDFT SLGTKISDEN FRHALIKGFL RNEYISNDLF | 480 |
| NKFWYDSKKY IEKILSDTSI IYKENCDKLL IVGIRWINVA DEFIREMKEN VKNVKNDNLS | 540 |
| DGKYLIFQII CEITYLIILY IRQHDDLCDK TLKLLDYILS KFTDIVFEDC RIVLELETFI | 600 |
| MEKIPKFITS LTKSMMSIFL IYITENKNIE NLKLLWISLF SHLSTIENKS KENIFVYEIA | 660 |
| QMIIDNKIDM KKLKGRLPQI DLIDHVLLNS VHLNIKDIFE NWESLNNLFA IKEIIFSKEV | 720 |
| IIESLLCLTE KVFFSNDSEF HSNIYDVKIL TLLYKVLNDS FDLYLCFLQN VRAIEFHIKM | 780 |
| WNFSQIEKEN ISENIVLFND MFEQVYCLSN KVGSDLRFNI ANIIQKQILS LSSLDFLLMT | 840 |
| SLHNLSIKIL FYVSEVEKAN LLNIFLFTSH EWNAAFTYLI FKYPNKNLEL FNSLKICCLI | 900 |
| FSNFIQTKKD IKVGNISDLL RMTVFSEMII YSLMLKSCIL EIHWLILYGI MVCGELVRDN | 960 |
| FELEHANCLW KSITEEQEDK LFALINRSKD LFFNILSSKK IKILGIDDVV EQNDIKFLKK | 1020 |
| VKERSIGKSP ETYYSACVLF RILQFLIEKN YYNQKEAEIW LNDINIENEQ NIFIIAAIVN | 1080 |
| GFQEKLFFSK KLENIRDRIC SDLSGKLVVD QDLKKLILFN IFIPFDDTLW RHLPQQRIET | 1140 |
| KIKNQPLLIC TQLLSEILTQ IPLNIIEKQL LYPHLLSQSR CTQIVTFDWL TNWISKNQED | 1200 |
| LILDTEFSKI NADLPSELLS MVLSPPYCYN LLNDIGHKNN MFCEIRCFFL SWILIFIHFK | 1260 |
| NTSFILKSSY IENLKNNNCI SIAIELIFKI LELLNNRFMN SIEILRDISK WKNIDDIKTE | 1320 |
| ILSLATYVYL LILKHVPSIA RDWWISCKNR QLTQLVETFT EKYISPILIE EEINLVLLEF | 1380 |
| TQLKITDNNI HVKISKTTRE VTTFYTIDDQ YLEMIIRLPS CYPLSQVIVE GLQKIGIKDA | 1440 |
| QWRAWLLASR ILVTTQNGSI VDAVNLFKRN VSLHFEGVAE CNICFSILSV QDRSLPSKKC | 1500 |
| ATCKNKFHSG CLYKWFKTSN ASKCPLYVFL VIWTNFLIAF FLKIFVWVSL LVGSNAYPLS | 1560 |
| IIRRGSRSIH NEGLSHSGNS QKGCLFPFNN FELSAVTSDN GKLGWATGLG DICSAGSYCS | 1620 |
| YYCRSGYHAR LNSRILGLSS SFSEGRLFCD GSGNLQMPSE SLCVRASHSV YLLNLFDVSL | 1680 |
| SFCERIFSNG GLRLASNSVN GRSFSELTIP YESSGPSSSF YYLGPPGTGR DGCTMGNRFS | 1740 |
| NQGLWSPYTL GISKDYKGDI HLTLGWNPTY IGDSYWSGVT PNWGIRIDCE GFGCGSKSCF | 1800 |
| IDPSVHGINE CENPVNSYSS AAFCAVRISL NTRTRVVLFS RSNVNDELKV LSSLPHFDFQ | 1860 |
| DSSETKLVPG TVDVSESKHE ESKSIVDKVE ITEHHESNDK VEDDTDAQVT VLNKQEAEES | 1920 |
| THTVDKNSDS EHSKTSELDS FDNKKSTFTD LLSKVDISSV TSLGQSSFGS NGYIFGNDGL | 1980 |
| TLMLFQLHGN KNNHIHTSGA SCPLSIHDNS NSLSVSSPHS VSDDCAPTNA LLFLDRHVSD | 2040 |
| GYKLLADKFD NYLTKISSLI RNGDFQVLEL DTPRNNNESD SVNNGTQHES SAYIMKNNIL | 2100 |
| WTSIEMYLLG HEVRNDITDV HYFKTISNRQ SFAIVYISVF VMTYCISLES QTTSNLIVPA | 2160 |
| TSYFGKNSLI PLINTINGVL YAVVKQQMAK LGVIFGRIEG FCFSLLLYDM GCMMYMMSTN | 2220 |
| IVTFIISSIL HTIGSTGIQI LQQIIISDMS NFLNRGLLNS ILDIPLLINV WVGPSLAQNI | 2280 |
| YLPEPASEQW RLGYGIWAVV LSIGSLPVII VLYLGQFRSE GIRMLSLENN DGSITFSMKR | 2340 |
| IFIELDLVGI MILSFGLIML LFQLIHEFPI FYDVKSLYII TIFVIVTILC VIFPLWECFY | 2400 |
| AKYPIFDITS FKLGMIGSGY FSSFFYFMGF YLYNNYFSTY LYVTKSNSIK RVGYLNNVFS | 2460 |
| FTSTIVAIFV GIFVKYNKRY NFFIYIGSPI YMIGIVYMIY NLRVDESVIE MTISQAIIGI | 2520 |
| SGGIYNMSAL IGIQTVSCRQ QVVVNTTLFL TLASLGGAIG SAISGIMWSS LLPKMLKIYS | 2580 |
| ERLNIFLTSS DYTLIIGSPF ILDQYPKFVK GTLGRMAIIL AYNDLMKVLY ISSAMCTIPM | 2640 |
| LFCVMFMKDI HLNRIKQEVY NDNVYKFSDE LKKH | 2674 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH132 | 1602 | 1830 | 9.6e-29 | 0.7491749174917491 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 340880 | MFS_ARN_like | 9.02e-108 | 2132 | 2649 | 1 | 513 | Yeast ARN family of Siderophore iron transporters and similar proteins of the Major Facilitator Superfamily. The ARN family of siderophore iron transporters includes ARN1 (or ferrichrome permease), ARN2 (or triacetylfusarinine C transporter 1 or TAF1), ARN3 (or siderophore iron transporter 1 or SIT1 or ferrioxamine B permease) and ARN4 (or Enterobactin permease or ENB1). They specifically recognize siderophore-iron chelates are expressed under conditions of iron deprivation. They facilitate the uptake of both hydroxamate- and catecholate-type siderophores. This group also includes glutathione exchanger 1 (Gex1p) and Gex2p, which are proton/glutathione antiporters that import glutathione from the vacuole and exports it through the plasma membrane. The ARN family belongs to the Major Facilitator Superfamily (MFS) of membrane transport proteins, which are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| 227544 | COG5219 | 2.68e-84 | 25 | 1526 | 7 | 1517 | Uncharacterized conserved protein, contains RING Zn-finger [General function prediction only]. |
| 397780 | SUN | 2.17e-30 | 1595 | 1805 | 17 | 233 | Beta-glucosidase (SUN family). Members of this family include Nca3, Sun4 and Sim1. This is a family of yeast proteins, involved in a diverse set of functions (DNA replication, aging, mitochondrial biogenesis and cell septation). BGLA from Candida wickerhamii has been characterized as a Beta-glucosidase EC:3.2.1.21. |
| 319405 | RING-CH-C4HC3_LTN1 | 1.05e-24 | 1480 | 1526 | 2 | 48 | RING-CH finger, H2 subclass (C4HC3-type), found in E3 ubiquitin-protein ligase listerin and similar proteins. Listerin, also known as RING finger protein 160 or zinc finger protein 294, is the mammalian homolog of yeast Ltn1. It is widely expressed in all tissues, but motor and sensory neurons and neuronal processes in the brainstem and spinal cord are primarily affected in the mutant. Listerin is required for embryonic development and plays an important role in neurodegeneration. It also functions as a critical E3 ligase involving quality control of nonstop proteins. It mediates ubiquitylation of aberrant proteins that become stalled on ribosomes during translation. Ltn1 works with several cofactors to form a large ribosomal subunit-associated quality control complex (RQC), whick mediates the ubiquitylation and extraction of ribosome-stalled nascent polypeptide chains for proteasomal degradation. It appears to first associate with nascent chain-stalled 60S subunits together with two proteins of unknown function, Tae2 and Rqc1. Listerin contains a long stretch of HEAT (Huntingtin, Elongation factor 3, PR65/A subunit of protein phosphatase 2A, and TOR) or ARM (Armadillo) repeats in the N terminus and middle region, and a catalytic RING-CH finger, also known as vRING or RINGv, with an unusual arrangement of zinc-coordinating residues in the C-terminus . Its cysteines and histidines are arranged in the sequence as C4HC3-type, rather than the C3H2C3-type in canonical RING-H2 finger. |
| 341045 | MFS_Azr1_MDR_like | 3.78e-14 | 2398 | 2563 | 151 | 315 | Saccharomyces cerevisiae Azole resistance protein 1 (Azr1p), and similar multidrug resistance (MDR) transporters of the Major Facilitator Superfamily. This subfamily is composed of multidrug resistance (MDR) transporters including various Saccharomyces cerevisiae proteins such as azole resistance protein 1 (Azr1p), vacuolar basic amino acid transporter 1 (Vba1p), vacuolar basic amino acid transporter 5 (Vba5p), and Sge1p (also known as Nor1p, 10-N-nonyl acridine orange resistance protein, and crystal violet resistance protein). MDR transporters are drug/H+ antiporters (DHA) that mediate the efflux of a variety of drugs and toxic compounds, and confer resistance to these compounds. This subfamily belongs to the Methylenomycin A resistance protein (also called MMR peptide) and similar multidrug resistance (MDR) transporters (MMR-like MDR transporter) family of the Major Facilitator Superfamily (MFS) of transporters. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QFZ39256.1|GH132 | 5.64e-35 | 1584 | 1840 | 23 | 282 |
| QFZ27107.1|GH132 | 5.64e-35 | 1584 | 1840 | 23 | 282 |
| QFZ50615.1|GH132 | 5.64e-35 | 1584 | 1840 | 23 | 282 |
| QFZ44938.1|GH132 | 5.64e-35 | 1584 | 1840 | 23 | 282 |
| QFZ33585.1|GH132 | 5.64e-35 | 1584 | 1840 | 23 | 282 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3J92_0 | 6.55e-52 | 59 | 1526 | 56 | 1760 | Structure and assembly pathway of the ribosome quality control complex [Homo sapiens],3J92_w Structure and assembly pathway of the ribosome quality control complex [Homo sapiens],3J92_z Structure and assembly pathway of the ribosome quality control complex [Homo sapiens] |
| 5FG0_A | 1.63e-23 | 51 | 351 | 11 | 324 | Structure of the conserved yeast listerin (Ltn1) N-terminal domain, MONOCLINIC FORM [Saccharomyces cerevisiae S288C],5FG0_B Structure of the conserved yeast listerin (Ltn1) N-terminal domain, MONOCLINIC FORM [Saccharomyces cerevisiae S288C] |
| 5FG1_A | 5.26e-23 | 51 | 351 | 11 | 324 | Structure of the conserved yeast listerin (Ltn1) selenomethionine-substituted N-terminal domain, TRIGONAL FORM [Saccharomyces cerevisiae S288C] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q4WHE1|MIRC_ASPFU | 1.57e-78 | 2130 | 2664 | 68 | 595 | MFS siderochrome iron transporter C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=mirC PE=2 SV=2 |
| sp|Q870L3|MIRC_EMENI | 3.99e-77 | 2130 | 2663 | 65 | 591 | Siderophore iron transporter mirC OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=mirC PE=3 SV=1 |
| sp|O74349|LTN1_SCHPO | 9.24e-66 | 52 | 1526 | 38 | 1602 | E3 ubiquitin-protein ligase listerin OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=rkr1 PE=3 SV=1 |
| sp|O74395|STR1_SCHPO | 1.30e-61 | 2117 | 2661 | 78 | 606 | Siderophore iron transporter 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=str1 PE=1 SV=1 |
| sp|Q04781|LTN1_YEAST | 2.36e-60 | 51 | 1526 | 21 | 1553 | E3 ubiquitin-protein ligase listerin OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=RKR1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000065 | 0.000001 |
TMHMM Annotations download full data without filtering help
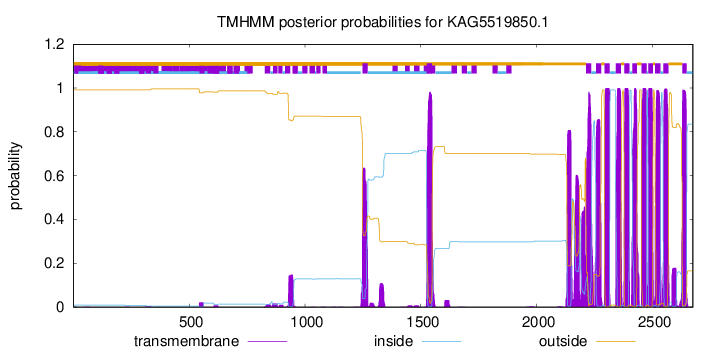
| Start | End |
|---|---|
| 1248 | 1270 |
| 1526 | 1548 |
| 2213 | 2235 |
| 2256 | 2278 |
| 2293 | 2315 |
| 2341 | 2363 |
| 2378 | 2400 |
| 2413 | 2432 |
| 2452 | 2474 |
| 2481 | 2501 |
| 2511 | 2533 |
| 2546 | 2568 |
| 2628 | 2647 |
