You are browsing environment: FUNGIDB
CAZyme Information: KAG5517874.1
You are here: Home > Sequence: KAG5517874.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pneumocystis sp. 'macacae' | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis sp. 'macacae' | |||||||||||
| CAZyme ID | KAG5517874.1 | |||||||||||
| CAZy Family | GH37 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT22 | 769 | 1131 | 6.4e-95 | 0.9562982005141388 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215437 | PLN02816 | 3.22e-85 | 770 | 1237 | 58 | 541 | mannosyltransferase |
| 281842 | Glyco_transf_22 | 1.29e-81 | 772 | 1131 | 20 | 414 | Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis. |
| 238121 | WD40 | 1.71e-63 | 364 | 631 | 5 | 258 | WD40 domain, found in a number of eukaryotic proteins that cover a wide variety of functions including adaptor/regulatory modules in signal transduction, pre-mRNA processing and cytoskeleton assembly; typically contains a GH dipeptide 11-24 residues from its N-terminus and the WD dipeptide at its C-terminus and is 40 residues long, hence the name WD40; between GH and WD lies a conserved core; serves as a stable propeller-like platform to which proteins can bind either stably or reversibly; forms a propeller-like structure with several blades where each blade is composed of a four-stranded anti-parallel b-sheet; instances with few detectable copies are hypothesized to form larger structures by dimerization; each WD40 sequence repeat forms the first three strands of one blade and the last strand in the next blade; the last C-terminal WD40 repeat completes the blade structure of the first WD40 repeat to create the closed ring propeller-structure; residues on the top and bottom surface of the propeller are proposed to coordinate interactions with other proteins and/or small ligands; 7 copies of the repeat are present in this alignment. |
| 238121 | WD40 | 1.90e-57 | 337 | 620 | 19 | 289 | WD40 domain, found in a number of eukaryotic proteins that cover a wide variety of functions including adaptor/regulatory modules in signal transduction, pre-mRNA processing and cytoskeleton assembly; typically contains a GH dipeptide 11-24 residues from its N-terminus and the WD dipeptide at its C-terminus and is 40 residues long, hence the name WD40; between GH and WD lies a conserved core; serves as a stable propeller-like platform to which proteins can bind either stably or reversibly; forms a propeller-like structure with several blades where each blade is composed of a four-stranded anti-parallel b-sheet; instances with few detectable copies are hypothesized to form larger structures by dimerization; each WD40 sequence repeat forms the first three strands of one blade and the last strand in the next blade; the last C-terminal WD40 repeat completes the blade structure of the first WD40 repeat to create the closed ring propeller-structure; residues on the top and bottom surface of the propeller are proposed to coordinate interactions with other proteins and/or small ligands; 7 copies of the repeat are present in this alignment. |
| 238121 | WD40 | 3.08e-56 | 182 | 577 | 1 | 288 | WD40 domain, found in a number of eukaryotic proteins that cover a wide variety of functions including adaptor/regulatory modules in signal transduction, pre-mRNA processing and cytoskeleton assembly; typically contains a GH dipeptide 11-24 residues from its N-terminus and the WD dipeptide at its C-terminus and is 40 residues long, hence the name WD40; between GH and WD lies a conserved core; serves as a stable propeller-like platform to which proteins can bind either stably or reversibly; forms a propeller-like structure with several blades where each blade is composed of a four-stranded anti-parallel b-sheet; instances with few detectable copies are hypothesized to form larger structures by dimerization; each WD40 sequence repeat forms the first three strands of one blade and the last strand in the next blade; the last C-terminal WD40 repeat completes the blade structure of the first WD40 repeat to create the closed ring propeller-structure; residues on the top and bottom surface of the propeller are proposed to coordinate interactions with other proteins and/or small ligands; 7 copies of the repeat are present in this alignment. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.79e-236 | 767 | 1232 | 27 | 493 | |
| 4.24e-116 | 767 | 1237 | 32 | 508 | |
| 5.90e-116 | 767 | 1237 | 32 | 508 | |
| 5.90e-116 | 767 | 1237 | 32 | 508 | |
| 5.27e-106 | 770 | 1239 | 62 | 548 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.37e-81 | 10 | 773 | 3 | 802 | Chain BC, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],5WYK_BC Chain BC, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6KE6_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6LQP_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6LQQ_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6LQR_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6LQS_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6LQT_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6LQU_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6LQV_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6ZQA_UM Chain UM, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6ZQB_UM Chain UM, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6ZQC_UM Chain UM, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6ZQD_UM Chain UM, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6ZQE_UM Chain UM, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],6ZQF_UM Chain UM, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],7AJT_UM Chain UM, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],7AJU_UM Chain UM, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],7D4I_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],7D5S_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],7D5T_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C],7D63_B3 Chain B3, U3 small nucleolar RNA-associated protein 13 [Saccharomyces cerevisiae S288C] |
|
| 3.28e-80 | 11 | 767 | 8 | 874 | Chain J, Utp13 [Thermochaetoides thermophila DSM 1495],6RXT_UM Chain UM, Utp13" [Thermochaetoides thermophila],6RXU_UM Chain UM, Utp13 [Thermochaetoides thermophila],6RXV_UM Chain UM, Utp13 [Thermochaetoides thermophila DSM 1495],6RXX_UM Chain UM, Utp13 [Thermochaetoides thermophila],6RXY_UM Chain UM, Utp13 [Thermochaetoides thermophila],6RXZ_UM Chain UM, Utp13 [Thermochaetoides thermophila] |
|
| 6.20e-66 | 11 | 770 | 12 | 774 | Chain LR, Transducin beta-like protein 3 [Homo sapiens],7MQ9_LR Chain LR, Transducin beta-like protein 3 [Homo sapiens],7MQA_LR Chain LR, Transducin beta-like protein 3 [Homo sapiens] |
|
| 8.49e-26 | 637 | 767 | 1 | 137 | Chain B, Structure of the CTD complex of UTP12, Utp13, Utp1 and Utp21 [Thermochaetoides thermophila DSM 1495] |
|
| 1.63e-25 | 637 | 767 | 1 | 137 | Chain B, Structure of the CTD complex of Utp12 and Utp13 [Thermochaetoides thermophila DSM 1495],5IC9_D Chain D, Structure of the CTD complex of Utp12 and Utp13 [Thermochaetoides thermophila DSM 1495] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.05e-116 | 767 | 1237 | 32 | 508 | GPI mannosyltransferase 3 OS=Yarrowia lipolytica (strain CLIB 122 / E 150) OX=284591 GN=GPI10 PE=3 SV=1 |
|
| 1.03e-114 | 11 | 768 | 7 | 764 | Probable U3 small nucleolar RNA-associated protein 13 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=utp13 PE=3 SV=3 |
|
| 9.36e-107 | 770 | 1239 | 62 | 548 | GPI mannosyltransferase 3 OS=Debaryomyces hansenii (strain ATCC 36239 / CBS 767 / BCRC 21394 / JCM 1990 / NBRC 0083 / IGC 2968) OX=284592 GN=GPI10 PE=3 SV=2 |
|
| 1.16e-91 | 770 | 1239 | 29 | 481 | GPI mannosyltransferase 3 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=GPI10 PE=3 SV=2 |
|
| 7.07e-81 | 10 | 773 | 3 | 802 | U3 small nucleolar RNA-associated protein 13 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=UTP13 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000047 | 0.000020 |
TMHMM Annotations download full data without filtering help
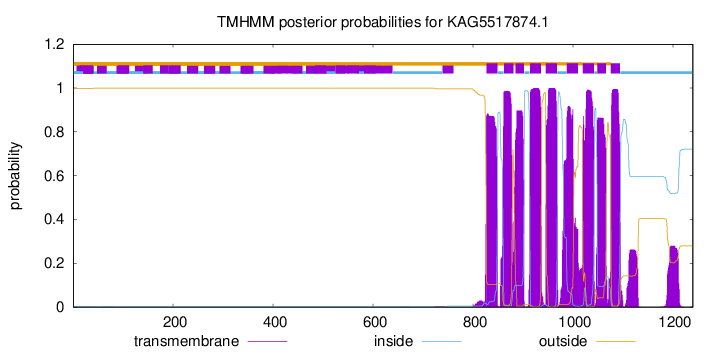
| Start | End |
|---|---|
| 827 | 849 |
| 862 | 881 |
| 885 | 902 |
| 914 | 936 |
| 946 | 968 |
| 988 | 1010 |
| 1020 | 1042 |
| 1049 | 1066 |
| 1076 | 1094 |
