You are browsing environment: FUNGIDB
CAZyme Information: KAG5438427.1
You are here: Home > Sequence: KAG5438427.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pneumocystis canis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis canis | |||||||||||
| CAZyme ID | KAG5438427.1 | |||||||||||
| CAZy Family | GT22 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH16 | 672 | 997 | 1.9e-149 | 0.9969325153374233 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397841 | SKN1 | 0.0 | 560 | 1047 | 2 | 500 | Beta-glucan synthesis-associated protein (SKN1). This family consists of the beta-glucan synthesis-associated proteins KRE6 and SKN1. Beta1,6-Glucan is a key component of the yeast cell wall, interconnecting cell wall proteins, beta1,3-glucan, and chitin. It has been postulated that the synthesis of beta1,6-glucan begins in the endoplasmic reticulum with the formation of protein-bound primer structures and that these primer structures are extended in the Golgi complex by two putative glucosyltransferases that are functionally redundant, Kre6 and Skn1. This is followed by maturation steps at the cell surface and by coupling to other cell wall macromolecules. |
| 395233 | Ribosomal_L3 | 0.0 | 1 | 366 | 1 | 369 | Ribosomal protein L3. |
| 240267 | PTZ00103 | 0.0 | 1 | 380 | 1 | 379 | 60S ribosomal protein L3; Provisional |
| 185689 | GH16_fungal_KRE6_glucanase | 7.96e-170 | 668 | 998 | 1 | 295 | Saccharomyces cerevisiae KRE6 and related glucanses, member of glycosyl hydrolase family 16. KRE6 is a Saccharomyces cerevisiae glucanase that participates in the synthesis of beta-1,6-glucan, a major structural component of the cell wall. It is a golgi membrane protein required for normal beta-1,6-glucan levels in the cell wall. KRE6 is closely realted to laminarinase, a glycosyl hydrolase family 16 member that hydrolyzes 1,3-beta-D-glucosidic linkages in 1,3-beta-D-glucans such as laminarins, curdlans, paramylons, and pachymans, with very limited action on mixed-link (1,3-1,4-)-beta-D-glucans. |
| 235260 | rpl3p | 1.18e-112 | 3 | 366 | 1 | 337 | 50S ribosomal protein L3P; Reviewed |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 0.0 | 382 | 1047 | 13 | 675 | |
| 0.0 | 378 | 1047 | 9 | 651 | |
| 2.06e-196 | 437 | 1047 | 96 | 700 | |
| 1.80e-192 | 542 | 1047 | 144 | 661 | |
| 1.83e-187 | 542 | 1047 | 140 | 657 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.45e-219 | 1 | 380 | 1 | 381 | Chain LB, 60S ribosomal protein L3-like protein [Thermochaetoides thermophila DSM 1495],7OLD_LB Chain LB, 60S ribosomal protein L3-like protein [Thermochaetoides thermophila DSM 1495] |
|
| 4.47e-212 | 1 | 380 | 1 | 380 | Chain AB, 60S ribosomal protein L3 [Kluyveromyces lactis] |
|
| 1.52e-211 | 1 | 379 | 1 | 380 | Chain E1, 60S ribosomal protein L3 [Neurospora crassa] |
|
| 2.26e-211 | 2 | 380 | 1 | 379 | Chain BB, 60S ribosomal protein L3 [Kluyveromyces lactis] |
|
| 2.28e-209 | 1 | 380 | 1 | 380 | Chain L3, 60S ribosomal protein L3 [Saccharomyces cerevisiae W303],3J6Y_L3 Chain L3, 60S ribosomal protein L3 [Saccharomyces cerevisiae W303],3J77_L3 Chain L3, 60S ribosomal protein L3 [Saccharomyces cerevisiae],3J78_L3 Chain L3, 60S ribosomal protein L3 [Saccharomyces cerevisiae],3JCT_B Cryo-em structure of eukaryotic pre-60S ribosomal subunits [Saccharomyces cerevisiae S288C],4V7F_C Arx1 pre-60S particle. [Saccharomyces cerevisiae],4V7R_BC Chain BC, 60S ribosomal protein L3 [Saccharomyces cerevisiae],4V7R_DC Chain DC, 60S ribosomal protein L3 [Saccharomyces cerevisiae],4V88_BB Chain BB, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],4V88_DB Chain DB, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],4V8T_B Cryo-EM Structure of the 60S Ribosomal Subunit in Complex with Arx1 and Rei1 [Saccharomyces cerevisiae S288C],4V91_B Kluyveromyces lactis 80S ribosome in complex with CrPV-IRES [Kluyveromyces lactis],5APN_B Structure of the yeast 60S ribosomal subunit in complex with Arx1, Alb1 and N-terminally tagged Rei1 [Saccharomyces cerevisiae S288C],5APO_B Structure of the yeast 60S ribosomal subunit in complex with Arx1, Alb1 and C-terminally tagged Rei1 [Saccharomyces cerevisiae S288C],5FL8_B CRYO-EM STRUCTURE OF THE RIX1-REA1 PRE-60S PARTICLE [Saccharomyces cerevisiae S288C],5GAK_F Yeast 60S ribosomal subunit with A-site tRNA, P-site tRNA and eIF-5A [Saccharomyces cerevisiae],5H4P_B Structural snapshot of cytoplasmic pre-60S ribosomal particles bound with Nmd3, Lsg1, Tif6 and Reh1 [Saccharomyces cerevisiae S288C],5JCS_B CRYO-EM STRUCTURE OF THE RIX1-REA1 PRE-60S PARTICLE [Saccharomyces cerevisiae S288C],5JUO_G Chain G, uL3 (yeast L3) [Saccharomyces cerevisiae],5JUP_G Chain G, uL3 (yeast L3) [Saccharomyces cerevisiae],5JUS_G Chain G, uL3 (yeast L3) [Saccharomyces cerevisiae],5JUT_G Chain G, uL3 (yeast L3) [Saccharomyces cerevisiae],5JUU_G Chain G, uL3 (yeast L3) [Saccharomyces cerevisiae],5MC6_BA Chain BA, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],5T62_E Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3-Tif6-Lsg1 Complex [Saccharomyces cerevisiae S288C],5T6R_E Nmd3 is a structural mimic of eIF5A, and activates the cpGTPase Lsg1 during 60S ribosome biogenesis: 60S-Nmd3 Complex [Saccharomyces cerevisiae S288C],5Z3G_F Cryo-EM structure of a nucleolar pre-60S ribosome (Rpf1-TAP) [Saccharomyces cerevisiae S288C],6C0F_B Yeast nucleolar pre-60S ribosomal subunit (state 2) [Saccharomyces cerevisiae BY4741],6ELZ_B State E (TAP-Flag-Ytm1 E80A) - Visualizing the assembly pathway of nucleolar pre-60S ribosmes [Saccharomyces cerevisiae S288C],6EM1_B State C (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes [Saccharomyces cerevisiae S288C],6EM4_B State B architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes [Saccharomyces cerevisiae S288C],6EM5_B State D architectural model (Nsa1-TAP Flag-Ytm1) - Visualizing the assembly pathway of nucleolar pre-60S ribosomes [Saccharomyces cerevisiae S288C],6FT6_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],6HD7_F Cryo-EM structure of the ribosome-NatA complex [Saccharomyces cerevisiae],6HHQ_CE Chain CE, 60S ribosomal protein L3 [Saccharomyces cerevisiae],6HHQ_k Chain k, 60S ribosomal protein L3 [Saccharomyces cerevisiae],6N8J_B Cryo-EM structure of late nuclear (LN) pre-60S ribosomal subunit [Saccharomyces cerevisiae S288C],6N8K_B Cryo-EM structure of early cytoplasmic-immediate (ECI) pre-60S ribosomal subunit [Saccharomyces cerevisiae S288C],6N8L_B Cryo-EM structure of early cytoplasmic-late (ECL) pre-60S ribosomal subunit [Saccharomyces cerevisiae S288C],6N8M_E Cryo-EM structure of pre-Lsg1 (PL) pre-60S ribosomal subunit [Saccharomyces cerevisiae S288C],6N8N_E Cryo-EM structure of Lsg1-engaged (LE) pre-60S ribosomal subunit [Saccharomyces cerevisiae S288C],6N8O_E Cryo-EM structure of Rpl10-inserted (RI) pre-60S ribosomal subunit [Saccharomyces cerevisiae S288C],6QIK_C Cryo-EM structures of Lsg1-TAP pre-60S ribosomal particles [Saccharomyces cerevisiae],6QT0_C Cryo-EM structures of Lsg1-TAP pre-60S ribosomal particles [Saccharomyces cerevisiae],6QTZ_C Cryo-EM structures of Lsg1-TAP pre-60S ribosomal particles [Saccharomyces cerevisiae],6RI5_C Cryo-EM structures of Lsg1-TAP pre-60S ribosomal particles [Saccharomyces cerevisiae],6RZZ_C Cryo-EM structures of Lsg1-TAP pre-60S ribosomal particles [Saccharomyces cerevisiae],6S05_C Cryo-EM structures of Lsg1-TAP pre-60S ribosomal particles [Saccharomyces cerevisiae],6SNT_i Chain i, 60S ribosomal protein L3 [Saccharomyces cerevisiae],6SV4_BA Chain BA, 60S ribosomal protein L3 [Saccharomyces cerevisiae],6SV4_YA Chain YA, 60S ribosomal protein L3 [Saccharomyces cerevisiae],6SV4_ZA Chain ZA, 60S ribosomal protein L3 [Saccharomyces cerevisiae],6T7I_LB Chain LB, 60S ribosomal protein L3 [Saccharomyces cerevisiae],6T83_By Chain By, 60S ribosomal protein L3 [Saccharomyces cerevisiae],6T83_Ea Chain Ea, 60S ribosomal protein L3 [Saccharomyces cerevisiae],6XIQ_B Chain B, RPL3 isoform 1 [Saccharomyces cerevisiae],6XIR_B Chain B, RPL3 isoform 1 [Saccharomyces cerevisiae],6YLG_B Rix1-Rea1 pre-60S particle - 60S core, body 1 (rigid body refinement) [Saccharomyces cerevisiae],6YLH_B Rix1-Rea1 pre-60S particle - full composite structure [Saccharomyces cerevisiae],6YLX_B pre-60S State NE1 (TAP-Flag-Nop53) [Saccharomyces cerevisiae],6YLY_B pre-60S State NE2 (TAP-Flag-Nop53) [Saccharomyces cerevisiae],6Z6J_LB Chain LB, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],6Z6K_LB Chain LB, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7AZY_l Chain l, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7BT6_B Cryo-EM structure of pre-60S ribosome from Saccharomyces cerevisiae rpl4delta63-87 strain at 3.12 Angstroms resolution(state R1) [Saccharomyces cerevisiae S288C],7BTB_B Cryo-EM structure of pre-60S ribosome from Saccharomyces cerevisiae rpl4delta63-87 strain at 3.22 Angstroms resolution(state R2) [Saccharomyces cerevisiae S288C],7OF1_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OH3_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OHP_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OHQ_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OHR_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OHS_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OHT_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OHU_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OHV_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OHW_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OHX_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OHY_B Chain B, 60S ribosomal protein L3 [Saccharomyces cerevisiae S288C],7OSA_uL3 Chain uL3, 60S ribosomal protein L3 [Saccharomyces cerevisiae],7OSM_uL3 Chain uL3, 60S ribosomal protein L3 [Saccharomyces cerevisiae],7RR5_LB Chain LB, RPL3 isoform 1 [Saccharomyces cerevisiae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.44e-229 | 1 | 380 | 1 | 380 | 60S ribosomal protein L3-B OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=rpl302 PE=3 SV=2 |
|
| 2.04e-229 | 1 | 380 | 1 | 380 | 60S ribosomal protein L3-A OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=rpl301 PE=1 SV=2 |
|
| 3.39e-215 | 1 | 380 | 1 | 381 | 60S ribosomal protein L3 OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=rpl3 PE=1 SV=2 |
|
| 1.02e-212 | 1 | 380 | 1 | 380 | 60S ribosomal protein L3 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=RPL3 PE=3 SV=1 |
|
| 2.30e-211 | 1 | 380 | 1 | 380 | 60S ribosomal protein L3 OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=RPL3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
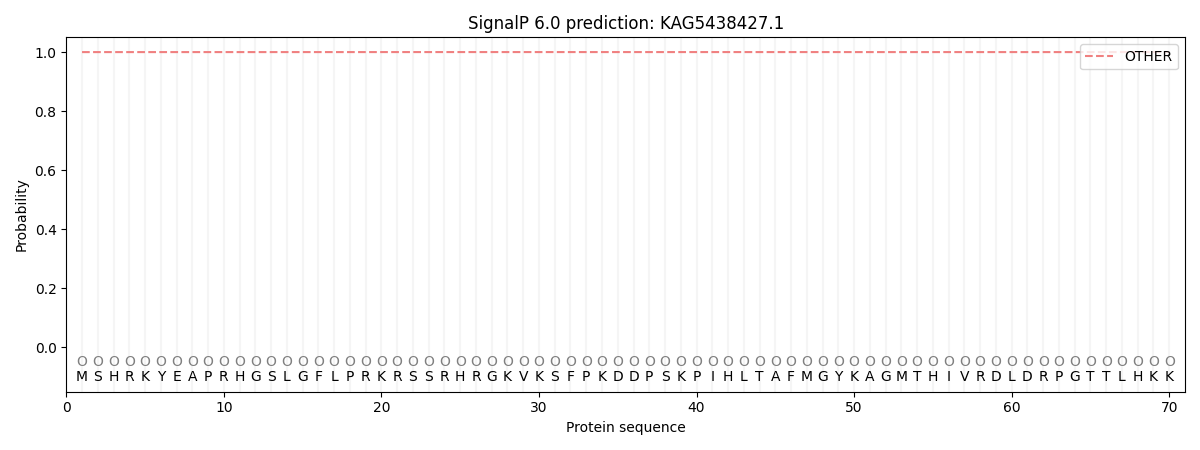
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000067 | 0.000000 |

