You are browsing environment: FUNGIDB
CAZyme Information: KAG5422039.1
You are here: Home > Sequence: KAG5422039.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Candida metapsilosis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; Candida metapsilosis | |||||||||||
| CAZyme ID | KAG5422039.1 | |||||||||||
| CAZy Family | GT71 | |||||||||||
| CAZyme Description | GLG1 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT8 | 77 | 382 | 1.9e-38 | 0.8715953307392996 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 133018 | GT8_Glycogenin | 8.85e-50 | 85 | 399 | 11 | 239 | Glycogenin belongs the GT 8 family and initiates the biosynthesis of glycogen. Glycogenin initiates the biosynthesis of glycogen by incorporating glucose residues through a self-glucosylation reaction at a Tyr residue, and then acts as substrate for chain elongation by glycogen synthase and branching enzyme. It contains a conserved DxD motif and an N-terminal beta-alpha-beta Rossmann-like fold that are common to the nucleotide-binding domains of most glycosyltransferases. The DxD motif is essential for coordination of the catalytic divalent cation, most commonly Mn2+. Glycogenin can be classified as a retaining glycosyltransferase, based on the relative anomeric stereochemistry of the substrate and product in the reaction catalyzed. It is placed in glycosyltransferase family 8 which includes lipopolysaccharide glucose and galactose transferases and galactinol synthases. |
| 396001 | Ribosomal_S21e | 5.30e-47 | 1 | 80 | 1 | 79 | Ribosomal protein S21e. |
| 132996 | Glyco_transf_8 | 1.34e-18 | 85 | 315 | 11 | 228 | Members of glycosyltransferase family 8 (GT-8) are involved in lipopolysaccharide biosynthesis and glycogen synthesis. Members of this family are involved in lipopolysaccharide biosynthesis and glycogen synthesis. GT-8 comprises enzymes with a number of known activities: lipopolysaccharide galactosyltransferase, lipopolysaccharide glucosyltransferase 1, glycogenin glucosyltransferase, and N-acetylglucosaminyltransferase. GT-8 enzymes contains a conserved DXD motif which is essential in the coordination of a catalytic divalent cation, most commonly Mn2+. |
| 279798 | Glyco_transf_8 | 3.07e-10 | 85 | 382 | 9 | 250 | Glycosyl transferase family 8. This family includes enzymes that transfer sugar residues to donor molecules. Members of this family are involved in lipopolysaccharide biosynthesis and glycogen synthesis. This family includes Lipopolysaccharide galactosyltransferase, lipopolysaccharide glucosyltransferase 1, and glycogenin glucosyltransferase. |
| 133037 | GT8_A4GalT_like | 1.73e-08 | 169 | 380 | 93 | 246 | A4GalT_like proteins catalyze the addition of galactose or glucose residues to the lipooligosaccharide (LOS) or lipopolysaccharide (LPS) of the bacterial cell surface. The members of this family of glycosyltransferases catalyze the addition of galactose or glucose residues to the lipooligosaccharide (LOS) or lipopolysaccharide (LPS) of the bacterial cell surface. The enzymes exhibit broad substrate specificities. The known functions found in this family include: Alpha-1,4-galactosyltransferase, LOS-alpha-1,3-D-galactosyltransferase, UDP-glucose:(galactosyl) LPS alpha1,2-glucosyltransferase, UDP-galactose: (glucosyl) LPS alpha1,2-galactosyltransferase, and UDP-glucose:(glucosyl) LPS alpha1,2-glucosyltransferase. Alpha-1,4-galactosyltransferase from N. meningitidis adds an alpha-galactose from UDP-Gal (the donor) to a terminal lactose (the acceptor) of the LOS structure of outer membrane. LOSs are virulence factors that enable the organism to evade the immune system of host cells. In E. coli, the three alpha-1,2-glycosyltransferases, that are involved in the synthesis of the outer core region of the LPS, are all members of this family. The three enzymes share 40 % of sequence identity, but have different sugar donor or acceptor specificities, representing the structural diversity of LPS. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.89e-317 | 84 | 671 | 12 | 605 | |
| 6.33e-309 | 84 | 671 | 12 | 609 | |
| 1.55e-184 | 84 | 671 | 12 | 660 | |
| 1.94e-181 | 84 | 671 | 12 | 666 | |
| 3.42e-107 | 85 | 671 | 13 | 579 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.50e-44 | 1 | 82 | 1 | 82 | Chain W, 40S ribosomal protein S21 [Candida albicans SC5314],7Q0F_W Chain W, 40S ribosomal protein S21 [Candida albicans SC5314],7Q0P_W Chain W, 40S ribosomal protein S21 [Candida albicans SC5314] |
|
| 1.91e-42 | 1 | 82 | 1 | 82 | CryoEM structure of 40S-eIF1-eIF1A preinitiation complex [Kluyveromyces lactis],3J81_V CryoEM structure of a partial yeast 48S preinitiation complex [Kluyveromyces lactis],3JAM_V CryoEM structure of 40S-eIF1A-eIF1 complex from yeast [Kluyveromyces lactis],3JAP_V Structure of a partial yeast 48S preinitiation complex in closed conformation [Kluyveromyces lactis],3JAQ_V Structure of a partial yeast 48S preinitiation complex in open conformation [Kluyveromyces lactis],5IT7_V Chain V, 40S ribosomal protein S21 [Kluyveromyces lactis],5IT9_V Structure of the yeast Kluyveromyces lactis small ribosomal subunit in complex with the cricket paralysis virus IRES. [Kluyveromyces lactis],6FYX_V Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C1) [Kluyveromyces lactis NRRL Y-1140],6FYY_V Structure of a partial yeast 48S preinitiation complex with eIF5 N-terminal domain (model C2) [Kluyveromyces lactis NRRL Y-1140],6GSM_V Chain V, 40S ribosomal protein S21 [Kluyveromyces lactis NRRL Y-1140],6GSN_V Chain V, 40S ribosomal protein S21 [Kluyveromyces lactis NRRL Y-1140],6UZ7_V Chain V, 40S ribosomal protein S21 [Kluyveromyces lactis] |
|
| 1.81e-41 | 1 | 82 | 1 | 82 | Chain 21, 40S ribosomal protein S21 [Saccharomyces cerevisiae W303],3J6Y_21 Chain 21, 40S ribosomal protein S21 [Saccharomyces cerevisiae W303],3J77_21 Chain 21, 40S ribosomal protein S21 [Saccharomyces cerevisiae],3J78_21 Chain 21, 40S ribosomal protein S21 [Saccharomyces cerevisiae],4U3M_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U3M_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U3N_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U3N_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U3U_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U3U_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4N_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4N_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4O_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4O_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4Q_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4Q_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4R_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4R_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4U_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4U_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4Y_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4Y_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4Z_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U4Z_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U50_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U50_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U51_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U51_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U52_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U52_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U53_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U53_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U55_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U55_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U56_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U56_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U6F_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4U6F_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4UER_Z 40S-eIF1-eIF1A-eIF3-eIF3j translation initiation complex from Lachancea kluyveri [Lachancea kluyveri],4V6I_AT Chain AT, 40S ribosomal protein rpS21 (S21e) [Saccharomyces cerevisiae],4V88_AV Chain AV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4V88_CV Chain CV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],4V8Y_AV Chain AV, 40S RIBOSOMAL PROTEIN S21-A [Saccharomyces cerevisiae],4V8Z_AV Chain AV, 40S RIBOSOMAL PROTEIN S21-A [Saccharomyces cerevisiae],4V92_BV Chain BV, ES21 [Kluyveromyces lactis],5DAT_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5DAT_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5DC3_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5DC3_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5DGE_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5DGE_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5DGF_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5DGF_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5DGV_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5DGV_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5FCI_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5FCI_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5FCJ_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5FCJ_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5I4L_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5I4L_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5JUO_SB Chain SB, eS21 (yeast S21) [Saccharomyces cerevisiae],5JUP_SB Chain SB, eS21 (yeast S21) [Saccharomyces cerevisiae],5JUS_SB Chain SB, eS21 (yeast S21) [Saccharomyces cerevisiae],5JUT_SB Chain SB, eS21 (yeast S21) [Saccharomyces cerevisiae],5JUU_SB Chain SB, eS21 (yeast S21) [Saccharomyces cerevisiae],5LL6_a Structure of the 40S ABCE1 post-splitting complex in ribosome recycling and translation initiation [Saccharomyces cerevisiae],5LYB_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],5LYB_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],5M1J_V2 Chain V2, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],5MC6_a Chain a, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5MEI_W Chain W, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5MEI_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5NDG_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],5NDG_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],5NDV_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5NDV_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5NDW_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5NDW_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5OBM_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5OBM_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5ON6_W Chain W, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5ON6_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5TBW_W Chain W, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5TBW_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],5TGA_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],5TGA_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],5TGM_D1 Chain D1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],5TGM_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6EML_a Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae [Saccharomyces cerevisiae S288C],6FAI_V Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit [Saccharomyces cerevisiae S288C],6GQ1_AL Chain AL, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],6GQB_AL Chain AL, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],6GQV_AL Chain AL, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6HHQ_W Chain W, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6HHQ_d1 Chain d1, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6I7O_a Chain a, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6I7O_ab Chain ab, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6Q8Y_a Chain a, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6RBD_V State 1 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles [Saccharomyces cerevisiae S288C],6RBE_V State 2 of yeast Tsr1-TAP Rps20-Deltaloop pre-40S particles [Saccharomyces cerevisiae S288C],6S47_BW Chain BW, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6SNT_V Chain V, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6SV4_a Chain a, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6SV4_ab Chain ab, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6SV4_ac Chain ac, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6T4Q_SV Chain SV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6T7I_SV Chain SV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6T7T_SV Chain SV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],6T83_Vb Chain Vb, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6T83_w Chain w, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6TB3_a Chain a, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],6TNU_a Chain a, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6WDR_V Subunit joining exposes nascent pre-40S rRNA for processing and quality control [Saccharomyces cerevisiae S288C],6WOO_VV Chain VV, eS21 [Saccharomyces cerevisiae],6XIQ_AL Chain AL, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6XIR_AL Chain AL, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],6Y7C_V Early cytoplasmic yeast pre-40S particle (purified with Tsr1 as bait) [Saccharomyces cerevisiae S288C],6Z6J_SV Chain SV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],6Z6K_SV Chain SV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],6ZCE_W Structure of a yeast ABCE1-bound 43S pre-initiation complex [Saccharomyces cerevisiae S288C],6ZU9_a Structure of a yeast ABCE1-bound 48S initiation complex [Saccharomyces cerevisiae S288C],6ZVI_D Mbf1-ribosome complex [Saccharomyces cerevisiae],7B7D_a Chain a, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],7MPI_BV Chain BV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],7MPJ_BV Chain BV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],7N8B_BV Chain BV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae],7NRC_Sa Chain Sa, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],7NRD_Sa Chain Sa, 40S ribosomal protein S21-A [Saccharomyces cerevisiae S288C],7OSA_eS21 Chain eS21, 40S ribosomal protein S21 [Saccharomyces cerevisiae],7OSM_eS21 Chain eS21, 40S ribosomal protein S21 [Saccharomyces cerevisiae],7RR5_SV Chain SV, 40S ribosomal protein S21-A [Saccharomyces cerevisiae] |
|
| 1.67e-35 | 1 | 82 | 1 | 82 | Chain SV, 40S ribosomal protein S21-like protein [Thermochaetoides thermophila DSM 1495],7OLD_SV Chain SV, 40S ribosomal protein S21-like protein [Thermochaetoides thermophila DSM 1495] |
|
| 4.26e-35 | 1 | 82 | 1 | 82 | Chain W2, 40S ribosomal protein S21 [Neurospora crassa] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7.71e-44 | 1 | 82 | 1 | 82 | 40S ribosomal protein S21 OS=Candida albicans OX=5476 GN=RPS21 PE=3 SV=1 |
|
| 9.81e-42 | 1 | 82 | 1 | 82 | 40S ribosomal protein S21 OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=RPS21 PE=1 SV=1 |
|
| 6.77e-41 | 1 | 82 | 1 | 82 | 40S ribosomal protein S21-B OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=RPS21B PE=1 SV=1 |
|
| 9.33e-41 | 1 | 82 | 1 | 82 | 40S ribosomal protein S21-A OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=RPS21A PE=1 SV=1 |
|
| 1.22e-39 | 1 | 82 | 1 | 82 | 40S ribosomal protein S21 OS=Candida glabrata (strain ATCC 2001 / CBS 138 / JCM 3761 / NBRC 0622 / NRRL Y-65) OX=284593 GN=RPS21 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
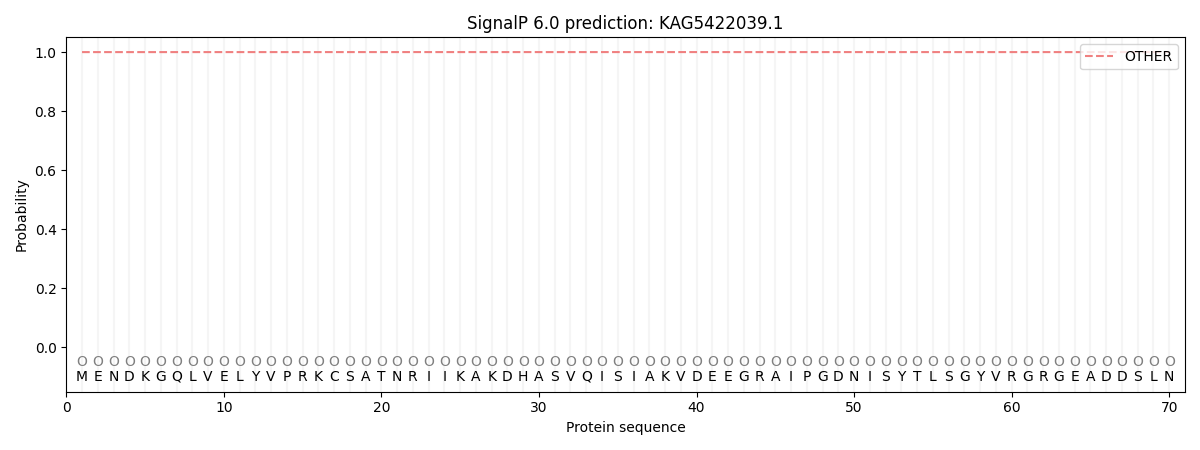
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000046 | 0.000000 |
