You are browsing environment: FUNGIDB
CAZyme Information: KAG5420226.1
You are here: Home > Sequence: KAG5420226.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Candida metapsilosis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Saccharomycetes; ; Debaryomycetaceae; Candida; Candida metapsilosis | |||||||||||
| CAZyme ID | KAG5420226.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | SCW11 | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH17 | 232 | 468 | 5e-21 | 0.909967845659164 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 227625 | Scw11 | 1.92e-68 | 208 | 468 | 43 | 302 | Exo-beta-1,3-glucanase, GH17 family [Carbohydrate transport and metabolism]. |
| 237874 | PRK14971 | 1.02e-04 | 72 | 143 | 384 | 455 | DNA polymerase III subunit gamma/tau. |
| 273167 | rad23 | 0.009 | 88 | 143 | 81 | 142 | UV excision repair protein Rad23. All proteins in this family for which functions are known are components of a multiprotein complex used for targeting nucleotide excision repair to specific parts of the genome. In humans, Rad23 complexes with the XPC protein. This family is based on the phylogenomic analysis of JA Eisen (1999, Ph.D. Thesis, Stanford University). [DNA metabolism, DNA replication, recombination, and repair] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 3.69e-213 | 1 | 471 | 1 | 463 | |
| 1.10e-212 | 1 | 471 | 1 | 474 | |
| 5.00e-148 | 11 | 471 | 12 | 489 | |
| 3.53e-139 | 11 | 471 | 12 | 506 | |
| 1.42e-134 | 202 | 471 | 55 | 324 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.38e-11 | 220 | 467 | 45 | 282 | Crystal structure of glycoside hydrolase family 17 beta-1,3-glucanosyltransferase from Rhizomucor miehei [Rhizomucor miehei CAU432] |
|
| 2.54e-10 | 220 | 467 | 45 | 282 | Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaribiose [Rhizomucor miehei CAU432],4WTS_A Active-site mutant of Rhizomucor miehei beta-1,3-glucanosyltransferase in complex with laminaritriose [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.16e-85 | 210 | 470 | 281 | 541 | Probable family 17 glucosidase SCW11 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW11 PE=1 SV=1 |
|
| 1.15e-50 | 213 | 470 | 311 | 564 | Probable beta-glucosidase btgE OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=btgE PE=3 SV=1 |
|
| 1.39e-48 | 213 | 470 | 305 | 558 | Probable beta-glucosidase btgE OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=btgE PE=3 SV=1 |
|
| 7.21e-48 | 213 | 468 | 139 | 387 | Probable family 17 glucosidase SCW10 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SCW10 PE=1 SV=1 |
|
| 1.36e-47 | 213 | 471 | 349 | 602 | Probable beta-glucosidase btgE OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=btgE PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
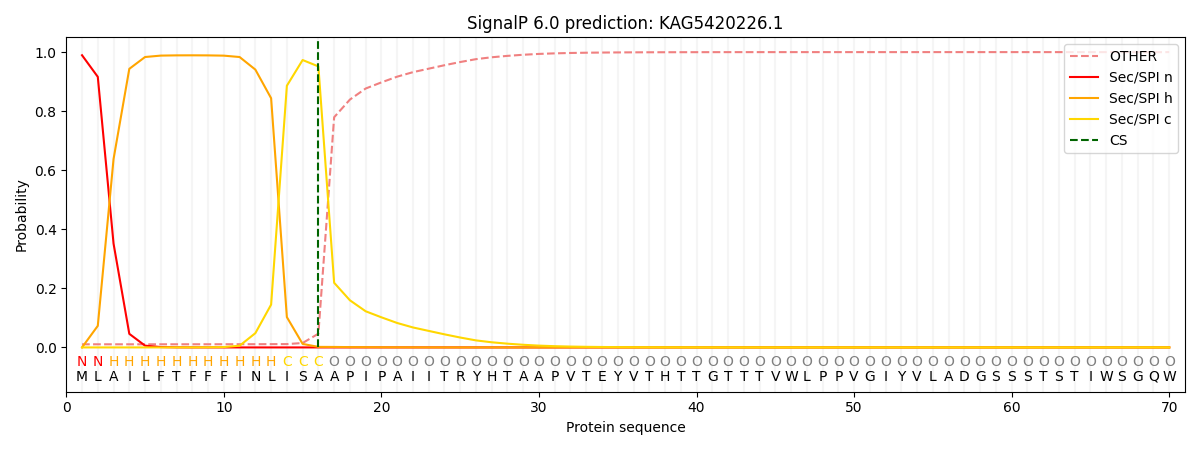
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.013452 | 0.986500 | CS pos: 16-17. Pr: 0.9516 |
