You are browsing environment: FUNGIDB
CAZyme Information: KAG4304593.1
You are here: Home > Sequence: KAG4304593.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
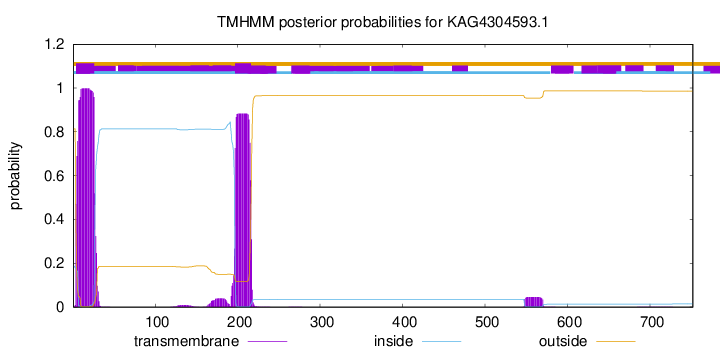
TMHMM annotations
Basic Information help
| Species | Pneumocystis oryctolagi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis oryctolagi | |||||||||||
| CAZyme ID | KAG4304593.1 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT32 | 75 | 156 | 2.4e-26 | 0.9555555555555556 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 226297 | OCH1 | 4.55e-64 | 43 | 264 | 64 | 290 | Mannosyltransferase OCH1 or related enzyme [Cell wall/membrane/envelope biogenesis]. |
| 398274 | Gly_transf_sug | 4.41e-28 | 73 | 158 | 1 | 90 | Glycosyltransferase sugar-binding region containing DXD motif. The DXD motif is a short conserved motif found in many families of glycosyltransferases, which add a range of different sugars to other sugars, phosphates and proteins. DXD-containing glycosyltransferases all use nucleoside diphosphate sugars as donors and require divalent cations, usually manganese. The DXD motif is expected to play a carbohydrate binding role in sugar-nucleoside diphosphate and manganese dependent glycosyltransferases. |
| 397779 | YjeF_N | 8.63e-27 | 519 | 701 | 2 | 168 | YjeF-related protein N-terminus. YjeF-N domain is a novel version of the Rossmann fold with a set of catalytic residues and structural features that are different from the conventional dehydrogenases. YjeF-N domain is fused to Ribokinases in bacteria (YjeF), where they may be phosphatases, and to divergent Sm and the FDF domain in eukaryotes (Dcp3p and FLJ21128), where they may be involved in decapping and catalyze hydrolytic RNA-processing reactions. |
| 223140 | Nnr1 | 5.61e-11 | 497 | 715 | 4 | 202 | NAD(P)H-hydrate repair enzyme Nnr, NAD(P)H-hydrate epimerase domain [Nucleotide transport and metabolism]. |
| 370547 | FDF | 2.11e-05 | 360 | 388 | 3 | 31 | FDF domain. The FDF domain, so called because of the conserved FDF at its N termini, is an entirely alpha-helical domain with multiple exposed hydrophilic loops. It is found at the C-terminus of Scd6p-like SM domains. It is also found with other divergent Sm domains and in proteins such as Dcp3p and FLJ21128, where it is found N terminal to the YjeF-N domain, a novel Rossmann fold domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.76e-126 | 1 | 265 | 1 | 265 | |
| 5.12e-80 | 20 | 265 | 20 | 266 | |
| 3.61e-79 | 13 | 265 | 13 | 266 | |
| 7.06e-79 | 13 | 265 | 13 | 266 | |
| 7.07e-79 | 13 | 265 | 13 | 266 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1.67e-68 | 57 | 269 | 60 | 274 | Mannosyl phosphorylinositol ceramide synthase CSH1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=CSH1 PE=1 SV=1 |
|
| 2.00e-67 | 55 | 269 | 51 | 266 | Mannosyl phosphorylinositol ceramide synthase SUR1 OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SUR1 PE=1 SV=1 |
|
| 5.05e-67 | 58 | 270 | 81 | 297 | Inositol phosphoceramide mannosyltransferase 2 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPCC4F11.04c PE=3 SV=1 |
|
| 5.12e-64 | 59 | 274 | 57 | 274 | Inositol phosphoceramide mannosyltransferase 3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPAC17G8.11c PE=1 SV=1 |
|
| 1.03e-51 | 57 | 274 | 62 | 281 | Inositol phosphoceramide mannosyltransferase 1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=imt1 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.256620 | 0.743354 | CS pos: 25-26. Pr: 0.5474 |