You are browsing environment: FUNGIDB
CAZyme Information: KAG4304114.1
You are here: Home > Sequence: KAG4304114.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pneumocystis oryctolagi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Ascomycota; Pneumocystidomycetes; ; Pneumocystidaceae; Pneumocystis; Pneumocystis oryctolagi | |||||||||||
| CAZyme ID | KAG4304114.1 | |||||||||||
| CAZy Family | GH16 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 2.4.1.-:14 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT50 | 153 | 389 | 2.1e-69 | 0.9198473282442748 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 215278 | PLN02502 | 0.0 | 644 | 1093 | 34 | 552 | lysyl-tRNA synthetase |
| 224111 | LysU | 5.20e-148 | 671 | 1092 | 12 | 500 | Lysyl-tRNA synthetase (class II) [Translation, ribosomal structure and biogenesis]. |
| 273107 | lysS_bact | 1.55e-138 | 672 | 1094 | 5 | 493 | lysyl-tRNA synthetase, eukaryotic and non-spirochete bacterial. This model represents the lysyl-tRNA synthetases that are class II amino-acyl tRNA synthetases. It includes all eukaryotic and most bacterial examples of the enzyme, but not archaeal or spirochete forms. [Protein synthesis, tRNA aminoacylation] |
| 234778 | lysS | 5.08e-138 | 671 | 1092 | 5 | 489 | lysyl-tRNA synthetase; Reviewed |
| 173607 | PTZ00417 | 5.94e-127 | 606 | 1092 | 18 | 583 | lysine-tRNA ligase; Provisional |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.00e-148 | 63 | 412 | 23 | 361 | |
| 3.86e-90 | 22 | 388 | 6 | 392 | |
| 3.86e-90 | 22 | 388 | 6 | 392 | |
| 4.33e-90 | 27 | 388 | 4 | 385 | |
| 1.60e-89 | 27 | 388 | 4 | 385 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.04e-169 | 671 | 1094 | 8 | 516 | Chain A, Lysine--tRNA ligase [Saccharomyces cerevisiae] |
|
| 5.96e-153 | 671 | 1093 | 9 | 511 | Loa loa Lysyl-tRNA synthetase in complex with Cladosporin. [Loa loa],5HGQ_B Loa loa Lysyl-tRNA synthetase in complex with Cladosporin. [Loa loa],5HGQ_C Loa loa Lysyl-tRNA synthetase in complex with Cladosporin. [Loa loa],5HGQ_D Loa loa Lysyl-tRNA synthetase in complex with Cladosporin. [Loa loa] |
|
| 8.17e-153 | 665 | 1093 | 2 | 508 | Crystal structure of cladosporin in complex with plasmodium like human lysyl-tRNA synthetase mutant [Homo sapiens],4YCW_B Crystal structure of cladosporin in complex with plasmodium like human lysyl-tRNA synthetase mutant [Homo sapiens],4YCW_E Crystal structure of cladosporin in complex with plasmodium like human lysyl-tRNA synthetase mutant [Homo sapiens],4YCW_F Crystal structure of cladosporin in complex with plasmodium like human lysyl-tRNA synthetase mutant [Homo sapiens] |
|
| 8.17e-153 | 665 | 1093 | 2 | 508 | Crystal Structure of Human LysRS: P38/AIMP2 Complex I [Homo sapiens],4DPG_B Crystal Structure of Human LysRS: P38/AIMP2 Complex I [Homo sapiens],4DPG_C Crystal Structure of Human LysRS: P38/AIMP2 Complex I [Homo sapiens],4DPG_D Crystal Structure of Human LysRS: P38/AIMP2 Complex I [Homo sapiens],4DPG_E Crystal Structure of Human LysRS: P38/AIMP2 Complex I [Homo sapiens],4DPG_F Crystal Structure of Human LysRS: P38/AIMP2 Complex I [Homo sapiens],4DPG_G Crystal Structure of Human LysRS: P38/AIMP2 Complex I [Homo sapiens],4DPG_H Crystal Structure of Human LysRS: P38/AIMP2 Complex I [Homo sapiens],4YCU_A Crystal structure of cladosporin in complex with human lysyl-tRNA synthetase [Homo sapiens],4YCU_B Crystal structure of cladosporin in complex with human lysyl-tRNA synthetase [Homo sapiens],6ILD_A Crystal Structure of Human LysRS: P38/AIMP2 Complex II [Homo sapiens],6ILD_B Crystal Structure of Human LysRS: P38/AIMP2 Complex II [Homo sapiens] |
|
| 1.07e-152 | 665 | 1093 | 2 | 508 | Crystal Structure of human lysyl-tRNA synthetase L350H mutant [Homo sapiens],6ILH_B Crystal Structure of human lysyl-tRNA synthetase L350H mutant [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5.14e-192 | 622 | 1094 | 10 | 581 | Lysine--tRNA ligase, cytoplasmic OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=krs1 PE=3 SV=1 |
|
| 4.83e-172 | 621 | 1094 | 10 | 580 | Lysine--tRNA ligase, cytoplasmic OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=KRS1 PE=1 SV=2 |
|
| 2.19e-163 | 628 | 1093 | 12 | 571 | Lysine--tRNA ligase cla4 OS=Cladosporium cladosporioides OX=29917 GN=cla4 PE=2 SV=1 |
|
| 3.95e-153 | 664 | 1093 | 69 | 576 | Lysine--tRNA ligase OS=Cricetulus griseus OX=10029 GN=KARS1 PE=1 SV=1 |
|
| 1.19e-151 | 664 | 1093 | 69 | 576 | Lysine--tRNA ligase OS=Homo sapiens OX=9606 GN=KARS1 PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 1.000008 | 0.000006 |
TMHMM Annotations download full data without filtering help
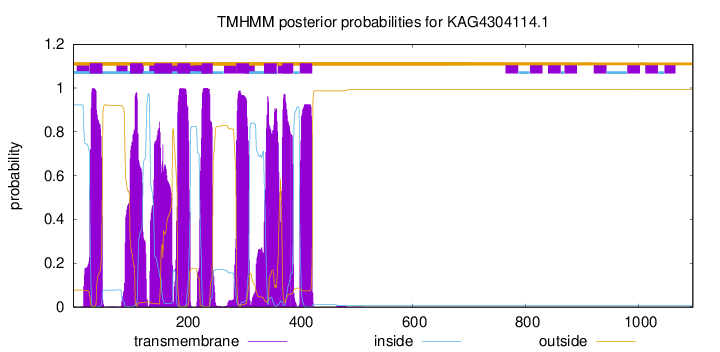
| Start | End |
|---|---|
| 30 | 52 |
| 101 | 123 |
| 144 | 175 |
| 185 | 207 |
| 228 | 247 |
| 289 | 311 |
| 338 | 360 |
| 370 | 389 |
| 401 | 423 |
