You are browsing environment: FUNGIDB
CAZyme Information: KAG2023521.1
Basic Information
help
| Species |
Coprinopsis cinerea
|
| Lineage |
Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea
|
| CAZyme ID |
KAG2023521.1
|
| CAZy Family |
PL8 |
| CAZyme Description |
unspecified product
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 588 |
|
67248.53 |
9.5125 |
|
| Genome Property |
| Genome Version/Assembly ID |
Genes |
Strain NCBI Taxon ID |
Non Protein Coding Genes |
Protein Coding Genes |
| FungiDB-61_Ccinerea326 |
15250 |
N/A |
238 |
15012
|
|
| Gene Location |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| 215596
|
PLN03133 |
3.51e-04 |
77 |
166 |
377 |
451 |
beta-1,3-galactosyltransferase; Provisional |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
| 1.45e-157 |
3 |
551 |
495 |
951 |
| 4.54e-122 |
7 |
554 |
158 |
788 |
| 2.26e-116 |
2 |
559 |
149 |
798 |
| 1.18e-115 |
2 |
552 |
149 |
793 |
| 2.39e-114 |
2 |
552 |
149 |
791 |
KAG2023521.1 has no PDB hit.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 2.24e-08 |
89 |
283 |
63 |
248 |
Beta-1,3-galactosyltransferase pvg3 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=pvg3 PE=1 SV=1 |
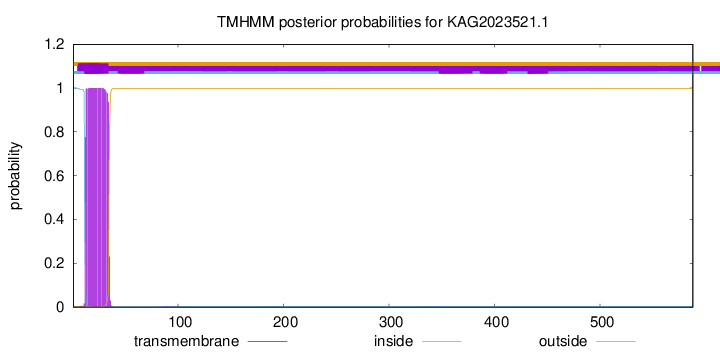
This protein is predicted as OTHER
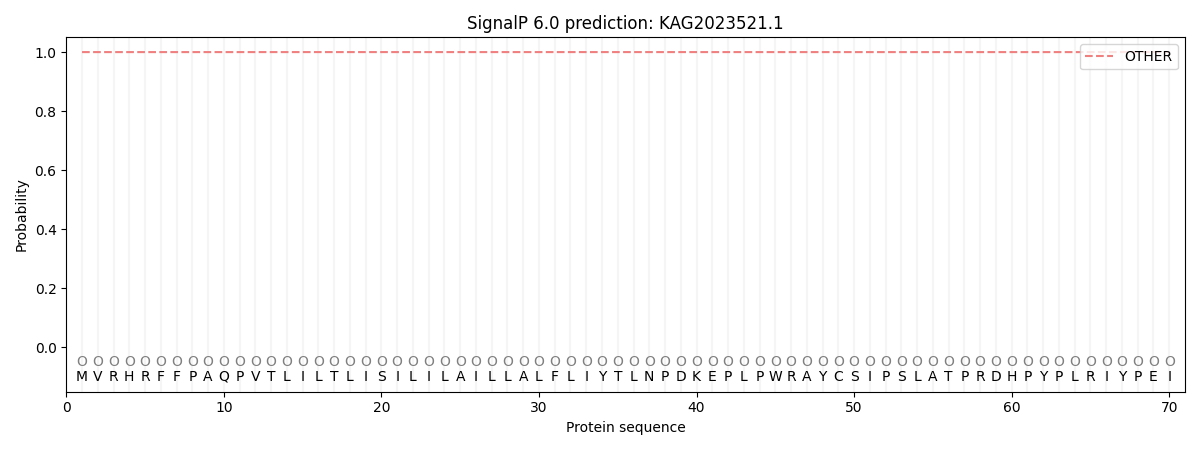
| Other |
SP_Sec_SPI |
CS Position |
| 1.000044 |
0.000001 |
|