You are browsing environment: FUNGIDB
CAZyme Information: KAG2022341.1
You are here: Home > Sequence: KAG2022341.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
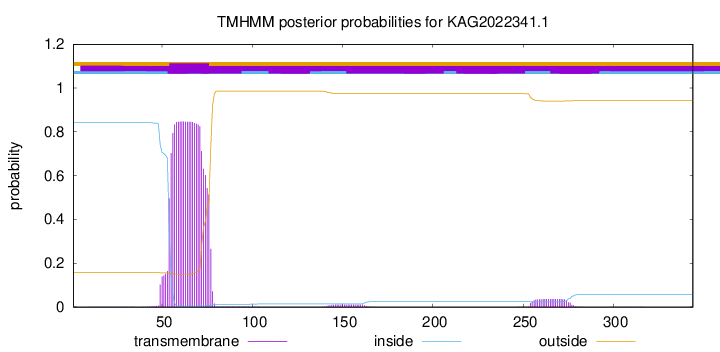
TMHMM annotations
Basic Information help
| Species | Coprinopsis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea | |||||||||||
| CAZyme ID | KAG2022341.1 | |||||||||||
| CAZy Family | GT48 | |||||||||||
| CAZyme Description | lipolytic enzyme | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE3 | 150 | 332 | 1.3e-59 | 0.9690721649484536 |
| CBM1 | 77 | 104 | 1.8e-16 | 0.9310344827586207 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 238871 | XynB_like | 8.23e-50 | 150 | 333 | 4 | 156 | SGNH_hydrolase subfamily, similar to Ruminococcus flavefaciens XynB. Most likely a secreted hydrolase with xylanase activity. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| 404371 | Lipase_GDSL_2 | 1.01e-23 | 152 | 325 | 2 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| 238141 | SGNH_hydrolase | 5.49e-18 | 152 | 332 | 4 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| 197593 | fCBD | 3.69e-16 | 75 | 108 | 1 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
| 395595 | CBM_1 | 5.74e-16 | 76 | 104 | 1 | 29 | Fungal cellulose binding domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 1.72e-51 | 143 | 343 | 31 | 234 | |
| 1.72e-51 | 143 | 343 | 31 | 234 | |
| 1.72e-51 | 143 | 343 | 31 | 234 | |
| 1.72e-51 | 143 | 343 | 31 | 234 | |
| 1.72e-51 | 143 | 343 | 31 | 234 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2.21e-28 | 151 | 331 | 10 | 192 | Chain A, LIPOLYTIC ENZYME [Acetivibrio thermocellus] |
|
| 2.57e-09 | 72 | 113 | 3 | 44 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor],6RV8_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor covalent complex with the aldouronic acid UXXR [Cerrena unicolor] |
|
| 3.89e-09 | 67 | 118 | 15 | 77 | Crystal structure of Endo-1,4-beta-xylanase-like protein from Acremonium chrysogenum [Acremonium chrysogenum ATCC 11550],5MRJ_B Crystal structure of Endo-1,4-beta-xylanase-like protein from Acremonium chrysogenum [Acremonium chrysogenum ATCC 11550] |
|
| 6.68e-09 | 152 | 327 | 7 | 192 | Crystal structure of a carbohydrate esterase family 3 from Talaromyces cellulolyticus [Talaromyces cellulolyticus] |
|
| 9.08e-09 | 62 | 107 | 539 | 584 | Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides],4QI5_A Chain A, Cellobiose dehydrogenase [Thermothelomyces myriococcoides] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.85e-15 | 145 | 330 | 42 | 256 | Multidomain esterase OS=Ruminococcus flavefaciens OX=1265 GN=cesA PE=1 SV=1 |
|
| 2.01e-14 | 66 | 111 | 10 | 56 | Putative endoglucanase type F OS=Fusarium oxysporum OX=5507 PE=2 SV=1 |
|
| 2.77e-13 | 58 | 116 | 3 | 66 | Acetylxylan esterase OS=Coprinopsis cinerea (strain Okayama-7 / 130 / ATCC MYA-4618 / FGSC 9003) OX=240176 GN=CC1G_12850 PE=1 SV=1 |
|
| 4.02e-13 | 59 | 114 | 4 | 58 | Endoglucanase gh5-1 OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=gh5-1 PE=1 SV=1 |
|
| 3.69e-12 | 72 | 116 | 20 | 65 | Exoglucanase 3 OS=Agaricus bisporus OX=5341 GN=cel3 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
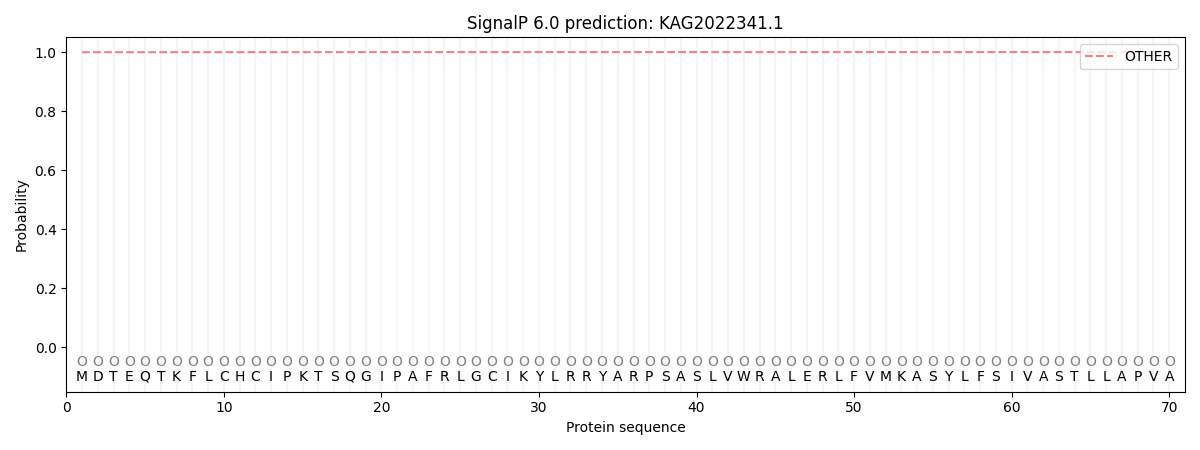
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.999709 | 0.000294 |