You are browsing environment: FUNGIDB
CAZyme Information: KAG2008967.1
You are here: Home > Sequence: KAG2008967.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprinopsis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea | |||||||||||
| CAZyme ID | KAG2008967.1 | |||||||||||
| CAZy Family | CE4 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.1.1.-:10 | 3.1.1.-:10 | 3.1.1.-:10 | 3.1.1.-:10 | 3.1.1.-:10 | 3.1.1.-:10 | 3.1.1.-:10 | 3.1.1.-:10 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE15 | 465 | 783 | 5.9e-80 | 0.9888475836431226 |
| CE15 | 2776 | 3098 | 6.4e-80 | 0.9888475836431226 |
| CE15 | 2388 | 2709 | 1.4e-79 | 0.9888475836431226 |
| CE15 | 80 | 402 | 2.3e-78 | 0.9888475836431226 |
| CE15 | 2002 | 2323 | 5.2e-78 | 0.9888475836431226 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 2.19e-123 | 2372 | 2743 | 2 | 376 | |
| 3.02e-123 | 459 | 816 | 110 | 468 | |
| 3.15e-119 | 464 | 816 | 110 | 459 | |
| 2.00e-118 | 464 | 816 | 110 | 459 | |
| 6.63e-118 | 463 | 816 | 108 | 458 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4.01e-121 | 464 | 816 | 25 | 374 | Chain A, Cip2 [Trichoderma reesei],3PIC_B Chain B, Cip2 [Trichoderma reesei],3PIC_C Chain C, Cip2 [Trichoderma reesei] |
|
| 4.30e-100 | 448 | 816 | 1 | 379 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor],6RU2_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor [Cerrena unicolor] |
|
| 6.06e-100 | 464 | 811 | 58 | 403 | Crystal structure of recombinant glucuronoyl esterase from Sporotrichum thermophile determined at 1.55 A resolution [Thermothelomyces thermophilus ATCC 42464] |
|
| 7.47e-100 | 446 | 816 | 5 | 385 | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant [Cerrena unicolor],6RTV_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant [Cerrena unicolor],6RU1_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X [Cerrena unicolor],6RU1_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid Um4X [Cerrena unicolor],6RV7_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR [Cerrena unicolor],6RV7_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR [Cerrena unicolor],6RV9_A Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR [Cerrena unicolor],6RV9_B Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid XUXXR [Cerrena unicolor] |
|
| 1.53e-99 | 464 | 811 | 58 | 403 | Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile determined at 1.9 A resolution [Thermothelomyces thermophilus ATCC 42464],4G4J_A Crystal structure of glucuronoyl esterase S213A mutant from Sporotrichum thermophile in complex with methyl 4-O-methyl-beta-D-glucopyranuronate determined at 2.35 A resolution [Thermothelomyces thermophilus ATCC 42464] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.55e-119 | 464 | 816 | 110 | 459 | 4-O-methyl-glucuronoyl methylesterase OS=Hypocrea jecorina (strain QM6a) OX=431241 GN=cip2 PE=1 SV=1 |
|
| 5.37e-111 | 433 | 816 | 96 | 480 | 4-O-methyl-glucuronoyl methylesterase OS=Podospora anserina (strain S / ATCC MYA-4624 / DSM 980 / FGSC 10383) OX=515849 GN=ge1 PE=1 SV=1 |
|
| 2.06e-105 | 465 | 816 | 36 | 392 | 4-O-methyl-glucuronoyl methylesterase OS=Schizophyllum commune (strain H4-8 / FGSC 9210) OX=578458 GN=SCHCODRAFT_238770 PE=1 SV=1 |
|
| 9.84e-100 | 464 | 811 | 44 | 389 | 4-O-methyl-glucuronoyl methylesterase OS=Myceliophthora thermophila (strain ATCC 42464 / BCRC 31852 / DSM 1799) OX=573729 GN=ge2 PE=1 SV=1 |
|
| 7.72e-99 | 459 | 816 | 37 | 393 | 4-O-methyl-glucuronoyl methylesterase OS=Neurospora crassa (strain ATCC 24698 / 74-OR23-1A / CBS 708.71 / DSM 1257 / FGSC 987) OX=367110 GN=Cip2 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
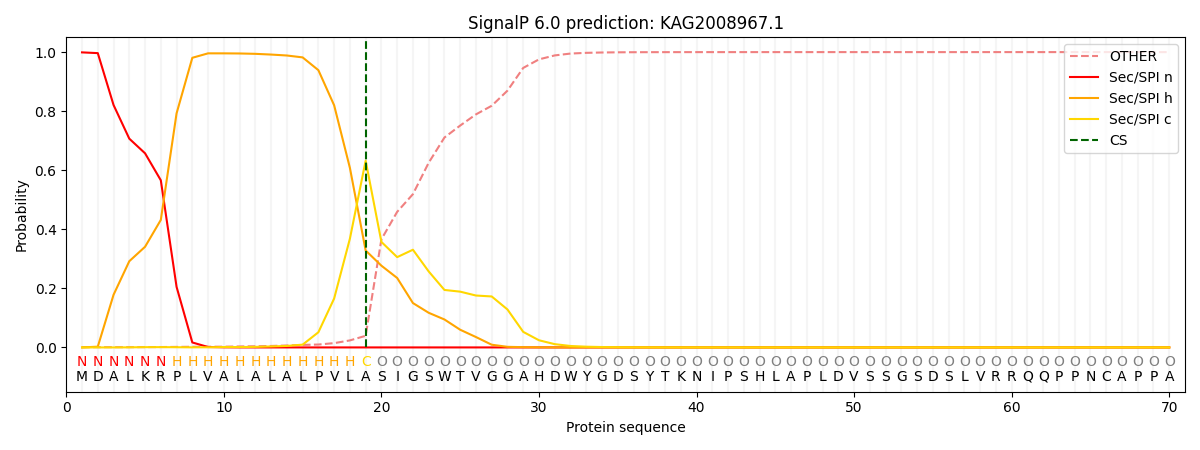
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.001645 | 0.998345 | CS pos: 19-20. Pr: 0.6325 |

