You are browsing environment: FUNGIDB
CAZyme Information: KAG2008178.1
You are here: Home > Sequence: KAG2008178.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprinopsis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea | |||||||||||
| CAZyme ID | KAG2008178.1 | |||||||||||
| CAZy Family | CBM21 | |||||||||||
| CAZyme Description | capsular associated protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 519208; End:523251 Strand: + | |||||||||||
Full Sequence Download help
| MVSFRLHRLP LRTVFFFILL VVILCTQISS WIPSFDVYTH LRPHVVEEER SCKVDNLVLT | 60 |
| RTATTTVFSP PQTVTVQEPV HTTESRDVFG FPVVGSREPS PSDAVDETRQ TARHFYRDDG | 120 |
| LLEVNPDGPH PIFELMEQAE QRWKKKNEKA SRTLKEACEE YERRYNRKPP RGFDVWWIYV | 180 |
| QSYDVRLPDE YDQISKNMEP FWGMDPRRLQ QIQRDWEDYT NTFTVGKETD TSPIEMLNHA | 240 |
| FDTEDGKSSR MTGAKMVADI LEEVSSFLPP FRAIFSSDDN PNLHTDYELM NMAIQAGKEG | 300 |
| RYIDIDNPPP VKLDGWISAC PPDSPAWVNH VEYTNPLPPL PELSRMKHNT TYASSPSPEA | 360 |
| LPPKTFIHNH KLSMGPCLHP AHLLMHGQFI SHERGPVPHR LIIPQFSFSP TALHHDITVA | 420 |
| LSINWIDDIR PRGHDPLFDD KDDERLQWRG SNTGSWFGGN TQWWVSHRTR LVDWANPRPL | 480 |
| QPSTIDSVLW SPPDSRWKVG EPSEPPSKSA WAPAMADIAF ANKPISCAPE VCDSRLAQEY | 540 |
| EFRHPHNIPT QGKYKYIIDV DGNAWSSRFK RLITSNSLIF KSTIYQEWFA DRIEPWLHYV | 600 |
| PIQIDYSDLL DALYFFRGDP GGRGAHPELA KKIAEAGREW SLTHWRKADL TAYMFRLFLE | 660 |
| YTRIMSPERE NGGLDYVYDE ADEFDPDVPV FGAEEEAEPR SGQSLFLRAM EKRRQHIVYP | 720 |
| NSFLTSKTPL EFTLDYLFGR FKPQPQVHVS HFKKITNGLL TCPPSSRSTS NASSNDDDAT | 780 |
| LNVLHLFYET WLSIYDDVRW FFLRDACSFT WYSTILNTTS TKKLSSSLNA TKTLLLILEK | 840 |
| LNTFPTEQSE LNAWWVPELG TKPPKPKQLR DGEDEDNASL SKPPGEEEEE EDNWRKFFED | 900 |
| DPTPKDPKDK QPSARLHQLT IHQSLHSLAS HRAVFTRTWL TLLPRVAASS SWKRRDEAST | 960 |
| FNRDLLTSKY RARFFRMANL FLASTHLPAT LLTSFVKRLA RLSLNAPPAA IVMIIPFTYN | 1020 |
| ILKRHPALMV MIHRDADDEE DPYSPTEPKP LSTNALSSSL WELYTHHSRI SWIIHTGLYS | 1080 |
| TRR | 1083 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 397822 | CBF | 9.58e-34 | 958 | 1067 | 26 | 143 | CBF/Mak21 family. |
| 214773 | CAP10 | 1.77e-21 | 551 | 665 | 143 | 253 | Putative lipopolysaccharide-modifying enzyme. |
| 310354 | Glyco_transf_90 | 1.75e-17 | 547 | 672 | 209 | 332 | Glycosyl transferase family 90. This family of glycosyl transferases are specifically (mannosyl) glucuronoxylomannan/galactoxylomannan -beta 1,2-xylosyltransferases, EC:2.4.2.-. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRW03676.1|GT90 | 5.61e-135 | 114 | 671 | 117 | 646 |
| QRV89460.1|GT90 | 5.61e-135 | 114 | 671 | 117 | 646 |
| QRV74770.1|GT90 | 5.61e-135 | 114 | 671 | 117 | 646 |
| CCA67199.1|GT90 | 6.21e-128 | 112 | 670 | 88 | 619 |
| CDR40950.1|GT90 | 1.74e-108 | 126 | 679 | 165 | 705 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6RXT_US | 2.02e-26 | 959 | 1063 | 345 | 460 | Chain US, Noc4 [Thermochaetoides thermophila],6RXU_US Chain US, Noc4 [Thermochaetoides thermophila],6RXV_US Chain US, Noc4 [Thermochaetoides thermophila DSM 1495],6RXX_US Chain US, Noc4 [Thermochaetoides thermophila],6RXY_US Chain US, Noc4 [Thermochaetoides thermophila],6RXZ_US Chain US, Noc4 [Thermochaetoides thermophila] |
| 7MQ8_SU | 7.56e-25 | 969 | 1063 | 298 | 394 | Chain SU, Nucleolar complex protein 4 homolog [Homo sapiens],7MQ9_SU Chain SU, Nucleolar complex protein 4 homolog [Homo sapiens],7MQA_SU Chain SU, Nucleolar complex protein 4 homolog [Homo sapiens] |
| 5WLC_SU | 3.32e-22 | 962 | 1063 | 346 | 466 | Chain SU, Noc4 [Saccharomyces cerevisiae BY4741] |
| 6KE6_RO | 3.32e-22 | 962 | 1063 | 346 | 466 | Chain RO, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6LQP_RO Chain RO, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6LQQ_RO Chain RO, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6LQR_RO Chain RO, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6LQS_RO Chain RO, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6LQU_RO Chain RO, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6LQV_RO Chain RO, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6ZQA_US Chain US, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6ZQB_US Chain US, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6ZQC_US Chain US, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6ZQD_US Chain US, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6ZQE_US Chain US, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6ZQF_US Chain US, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],6ZQG_US Chain US, Noc4,Nucleolar complex protein 4,Noc4 [Saccharomyces cerevisiae S288C],7AJT_US Chain US, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],7AJU_US Chain US, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],7D4I_RO Chain RO, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],7D5S_RO Chain RO, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C],7D63_RO Chain RO, Nucleolar complex protein 4 [Saccharomyces cerevisiae S288C] |
| 5L0R_A | 7.12e-14 | 553 | 666 | 236 | 343 | human POGLUT1 in complex with Notch1 EGF12 and UDP [Homo sapiens],5L0S_A human POGLUT1 in complex with Factor VII EGF1 and UDP [Homo sapiens],5L0T_A human POGLUT1 in complex with EGF(+) and UDP [Homo sapiens],5L0U_A human POGLUT1 in complex with EGF(+) and UDP-phosphono-glucose [Homo sapiens],5L0V_A human POGLUT1 in complex with 2F-glucose modified EGF(+) and UDP [Homo sapiens],5UB5_A human POGLUT1 in complex with human Notch1 EGF12 S458T mutant and UDP [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| sp|Q5K8R6|CXT1_CRYNJ | 3.83e-80 | 130 | 677 | 152 | 668 | Beta-1,2-xylosyltransferase 1 OS=Cryptococcus neoformans var. neoformans serotype D (strain JEC21 / ATCC MYA-565) OX=214684 GN=CXT1 PE=1 SV=1 |
| sp|O94372|YG06_SCHPO | 5.54e-27 | 962 | 1065 | 313 | 416 | Uncharacterized protein C1604.06c OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=SPBC1604.06c PE=3 SV=1 |
| sp|Q8BHY2|NOC4L_MOUSE | 1.86e-24 | 969 | 1071 | 342 | 450 | Nucleolar complex protein 4 homolog OS=Mus musculus OX=10090 GN=Noc4l PE=2 SV=1 |
| sp|Q5I0I8|NOC4L_RAT | 1.86e-24 | 969 | 1071 | 342 | 450 | Nucleolar complex protein 4 homolog OS=Rattus norvegicus OX=10116 GN=Noc4l PE=2 SV=1 |
| sp|Q4VBT2|NOC4L_DANRE | 4.75e-24 | 969 | 1071 | 351 | 460 | Nucleolar complex protein 4 homolog OS=Danio rerio OX=7955 GN=noc4l PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
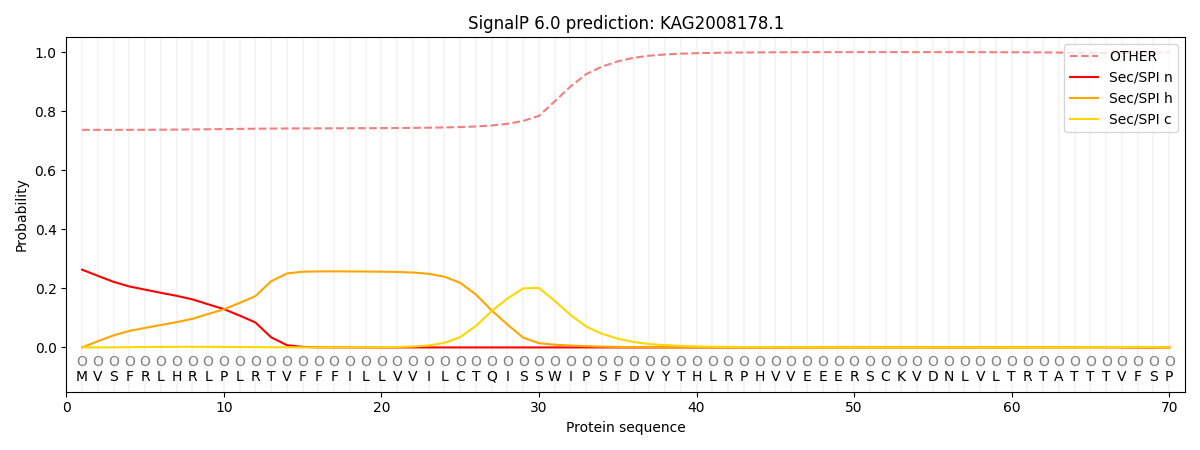
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.749374 | 0.250637 |

