You are browsing environment: FUNGIDB
CAZyme Information: KAG2006955.1
You are here: Home > Sequence: KAG2006955.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Coprinopsis cinerea | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea | |||||||||||
| CAZyme ID | KAG2006955.1 | |||||||||||
| CAZy Family | AA9 | |||||||||||
| CAZyme Description | endoglucanase II | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| AA12 | 40 | 436 | 1.2e-125 | 0.9950124688279302 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 225044 | YliI | 1.68e-13 | 42 | 382 | 39 | 374 | Glucose/arabinose dehydrogenase, beta-propeller fold [Carbohydrate transport and metabolism]. |
| 395595 | CBM_1 | 1.76e-10 | 478 | 506 | 1 | 29 | Fungal cellulose binding domain. |
| 197593 | fCBD | 5.38e-10 | 479 | 510 | 3 | 34 | Fungal-type cellulose-binding domain. Small four-cysteine cellulose-binding domain of fungi |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.44e-260 | 40 | 512 | 244 | 726 | |
| 4.44e-260 | 40 | 512 | 244 | 726 | |
| 1.58e-99 | 27 | 436 | 19 | 434 | |
| 2.17e-99 | 40 | 436 | 30 | 434 | |
| 7.47e-99 | 40 | 438 | 246 | 653 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3.38e-238 | 40 | 438 | 5 | 410 | Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_A Chain A, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea],6JWF_B Chain B, Extracellular PQQ-dependent sugar dehydrogenase [Coprinopsis cinerea] |
|
| 5.68e-67 | 45 | 438 | 5 | 402 | Iodide structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 5.83e-67 | 45 | 438 | 6 | 403 | Native structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 5.99e-67 | 45 | 438 | 7 | 404 | Calcium structure of Trichoderma reesei Carbohydrate-Active Enzymes Family AA12 [Trichoderma reesei RUT C-30] |
|
| 4.89e-09 | 475 | 510 | 1 | 36 | Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2CBH_A Chain A, C-TERMINAL DOMAIN OF CELLOBIOHYDROLASE I [Trichoderma reesei],2MWJ_A Chain A, Exoglucanase 1 [Trichoderma reesei],2MWK_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X34_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X35_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X36_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X37_A Chain A, Exoglucanase 1 [Trichoderma reesei],5X38_A Chain A, Exoglucanase 1 [Trichoderma reesei] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.13e-10 | 476 | 511 | 207 | 242 | Endoglucanase-5 OS=Hypocrea jecorina OX=51453 GN=egl5 PE=3 SV=1 |
|
| 1.01e-09 | 473 | 511 | 18 | 56 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=cbhC PE=3 SV=1 |
|
| 1.01e-09 | 473 | 511 | 18 | 56 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=cbhC PE=3 SV=1 |
|
| 1.04e-09 | 26 | 364 | 9 | 320 | Uncharacterized protein BB_0024 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0024 PE=4 SV=1 |
|
| 1.33e-09 | 473 | 511 | 18 | 56 | Probable 1,4-beta-D-glucan cellobiohydrolase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=cbhC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
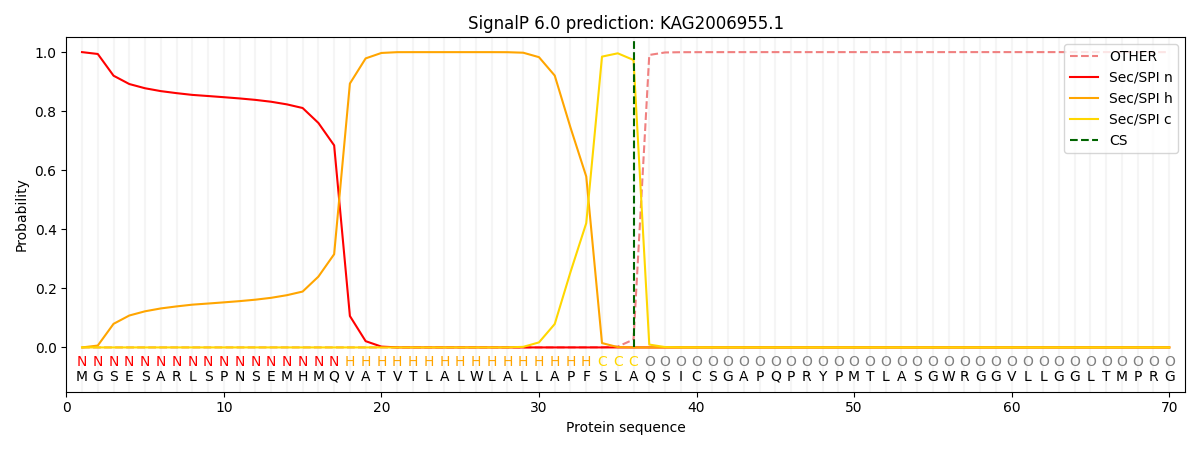
| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000241 | 0.999719 | CS pos: 36-37. Pr: 0.9733 |
