You are browsing environment: FUNGIDB
KAG2003241.1
Basic Information
help
Species
Coprinopsis cinerea
Lineage
Basidiomycota; Agaricomycetes; ; Psathyrellaceae; Coprinopsis; Coprinopsis cinerea
CAZyme ID
KAG2003241.1
CAZy Family
AA2
CAZyme Description
glycosyl hydrolase family 88
CAZyme Property
Genome Property
Genome Version/Assembly ID
Genes
Strain NCBI Taxon ID
Non Protein Coding Genes
Protein Coding Genes
FungiDB-61_Ccinerea326
15250
N/A
238
15012
Gene Location
No EC number prediction in KAG2003241.1.
Family
Start
End
Evalue
family coverage
GH105
49
317
6.1e-21
0.75
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
400034
Glyco_hydro_88
2.92e-12
53
362
29
295
Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases.
more
226678
YesR
8.26e-12
131
387
97
347
Rhamnogalacturonyl hydrolase YesR [Carbohydrate transport and metabolism].
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
2.66e-97
35
398
29
375
4.70e-97
35
398
34
380
1.64e-96
35
395
107
450
2.00e-96
35
398
34
380
3.89e-93
35
398
29
375
KAG2003241.1 has no PDB hit.
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
4.17e-07
131
330
91
280
Unsaturated rhamnogalacturonyl hydrolase YesR OS=Bacillus subtilis (strain 168) OX=224308 GN=yesR PE=1 SV=1
more
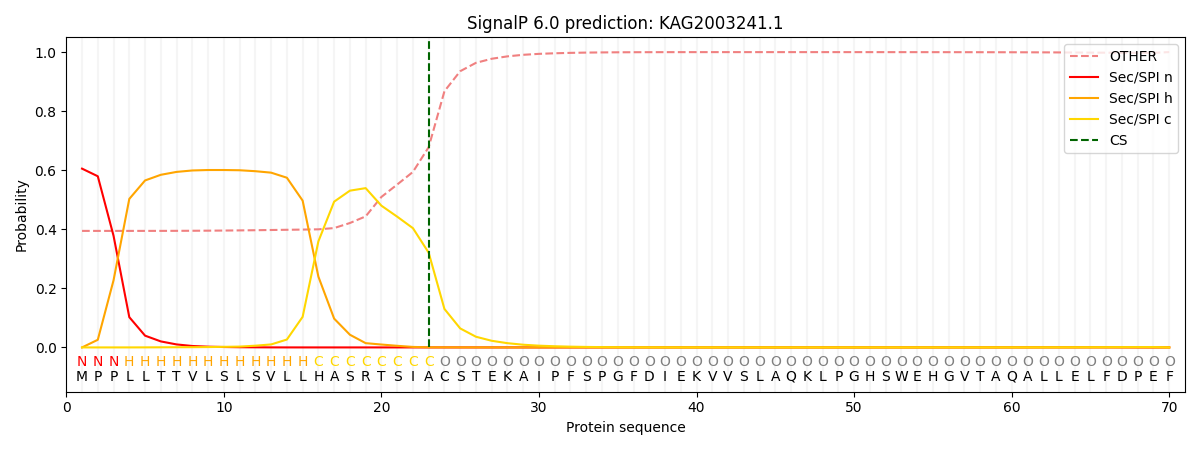
This protein is predicted as SP
Other
SP_Sec_SPI
CS Position
0.414072
0.585918
CS pos: 23-24. Pr: 0.3207
There is no transmembrane helices in KAG2003241.1.