You are browsing environment: FUNGIDB
CAZyme Information: KAG1712313.1
You are here: Home > Sequence: KAG1712313.1
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
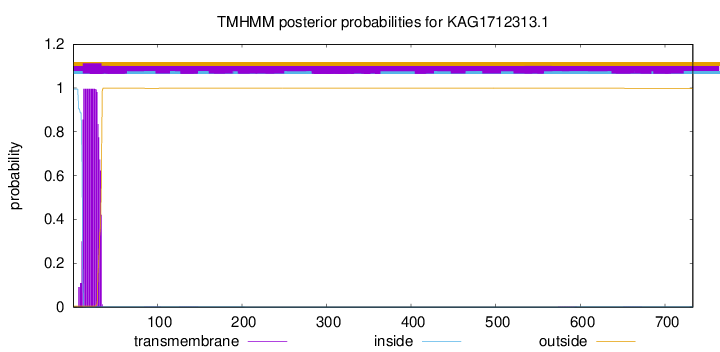
TMHMM annotations
Basic Information help
| Species | Phytophthora capsici | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Oomycota; NA; ; Peronosporaceae; Phytophthora; Phytophthora capsici | |||||||||||
| CAZyme ID | KAG1712313.1 | |||||||||||
| CAZy Family | GT8 | |||||||||||
| CAZyme Description | unspecified product | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | ||||||||||||
Enzyme Prediction help
| EC | 3.2.1.113:20 |
|---|
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH47 | 46 | 529 | 1.6e-135 | 0.9932735426008968 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| 396217 | Glyco_hydro_47 | 1.02e-137 | 46 | 529 | 1 | 451 | Glycosyl hydrolase family 47. Members of this family are alpha-mannosidases that catalyze the hydrolysis of the terminal 1,2-linked alpha-D-mannose residues in the oligo-mannose oligosaccharide Man(9)(GlcNAc)(2). |
| 240427 | PTZ00470 | 6.16e-73 | 35 | 478 | 71 | 503 | glycoside hydrolase family 47 protein; Provisional |
| 239041 | PA_EDEM3_like | 3.37e-18 | 595 | 716 | 2 | 121 | PA_EDEM3_like: protease associated domain (PA) domain-containing EDEM3-like proteins. This group contains various PA domain-containing proteins similar to mouse EDEM3 (ER-degradation-enhancing mannosidase-like 3 protein). EDEM3 contains a region, similar to Class I alpha-mannosidases (gylcosyl hydrolase family 47), N-terminal to the PA domain. EDEM3 accelerates glycoprotein ERAD (ER-associated degradation). In transfected mammalian cells, overexpression of EDEM3 enhances the mannose trimming from the N-glycans, of a model misfolded protein [alpha1-antitrypsin null (Hong Kong)] as well as, from total glycoproteins. Mannose trimming appears to be involved in the selection of ERAD substrates. EDEM3 has a different specificity of trimming than ER alpha-mannosidase 1. The significance of the PA domain to EDEM3 has not been ascertained. It may be a protein-protein interaction domain. At peptidase active sites, the PA domain may participate in substrate binding and/or promoting conformational changes, which influence the stability and accessibility of the site to substrate. |
| 240122 | PA_subtilisin_1 | 4.74e-16 | 597 | 707 | 2 | 104 | PA_subtilisin_1: Protease-associated domain containing subtilisin-like proteases, subgroup 1. A subgroup of PA domain-containing subtilisin-like proteases. The significance of the PA domain to many of the proteins in which it is inserted is undetermined. It may be a protein-protein interaction domain. At peptidase active sites, the PA domain may participate in substrate binding and/or promoting conformational changes, which influence the stability and accessibility of the site to substrate. Proteins into which the PA domain is inserted include the following subtilisin-like proteases: i) melon cucumisin, ii) Arabidopsis thaliana Ara12, iii) Alnus glutinosa ag12, iv) members of the tomato P69 family, and v) tomato LeSBT2. However, these proteins belong to other subtilisin-like subgroups. Relatively little is known about proteins in this subgroup. |
| 238300 | PA | 1.16e-10 | 622 | 709 | 30 | 114 | PA: Protease-associated (PA) domain. The PA domain is an insert domain in a diverse fraction of proteases. The significance of the PA domain to many of the proteins in which it is inserted is undetermined. It may be a protein-protein interaction domain. At peptidase active sites, the PA domain may participate in substrate binding and/or promoting conformational changes, which influence the stability and accessibility of the site to substrate. Proteins into which the PA domain is inserted include the following: i) various signal peptide peptidases including, hSPPL2a and 2b which catalyze the intramembrane proteolysis of tumor necrosis factor alpha, ii) various proteins containing a C3H2C3 RING finger including, Arabidopsis ReMembR-H2 protein and various E3 ubiquitin ligases such as human GRAIL (gene related to anergy in lymphocytes), iii) EDEM3 (ER-degradation-enhancing mannosidase-like 3 protein), iv) various plant vacuolar sorting receptors such as Pisum sativum BP-80, v) glutamate carboxypeptidase II (GCPII), vi) yeast aminopeptidase Y, vii) Vibrio metschnikovii VapT, a sodium dodecyl sulfate (SDS) resistant extracellular alkaline serine protease, viii) lactocepin (a cell envelope-associated protease from Lactobacillus paracasei subsp. paracasei NCDO 151), ix) various subtilisin-like proteases such as melon Cucumisin, and x) human TfR (transferrin receptor) 1 and 2. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| 4.36e-262 | 14 | 733 | 3 | 727 | |
| 2.89e-237 | 16 | 728 | 10 | 695 | |
| 3.63e-136 | 6 | 699 | 10 | 738 | |
| 6.51e-136 | 6 | 699 | 10 | 747 | |
| 6.51e-136 | 6 | 699 | 10 | 747 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 8.97e-60 | 40 | 476 | 6 | 435 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase and Man9GlcNAc2-PA complex [Homo sapiens] |
|
| 9.64e-60 | 40 | 476 | 6 | 435 | Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens],5KK7_B Crystal structure of the class I human endoplasmic reticulum 1,2-alpha-mannosidase T688A mutant and Thio-disaccharide substrate analog complex [Homo sapiens] |
|
| 1.04e-59 | 40 | 476 | 11 | 440 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase [Homo sapiens] |
|
| 1.09e-59 | 40 | 476 | 11 | 440 | Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With 1-Deoxymannojirimycin [Homo sapiens],1FO3_A Crystal Structure Of Human Class I Alpha1,2-Mannosidase In Complex With Kifunensine [Homo sapiens] |
|
| 6.17e-59 | 40 | 476 | 89 | 518 | Crystal Structure Of Human Class I alpha-1,2-Mannosidase In Complex With Thio-Disaccharide Substrate Analogue [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6.87e-123 | 36 | 475 | 122 | 563 | ER degradation-enhancing alpha-mannosidase-like protein 1 OS=Mus musculus OX=10090 GN=Edem1 PE=1 SV=1 |
|
| 1.57e-122 | 36 | 535 | 127 | 590 | ER degradation-enhancing alpha-mannosidase-like protein 1 OS=Homo sapiens OX=9606 GN=EDEM1 PE=1 SV=1 |
|
| 1.75e-122 | 14 | 699 | 14 | 754 | ER degradation-enhancing alpha-mannosidase-like protein 3 OS=Xenopus laevis OX=8355 GN=edem3 PE=2 SV=2 |
|
| 1.42e-121 | 11 | 713 | 29 | 788 | ER degradation-enhancing alpha-mannosidase-like protein 3 OS=Mus musculus OX=10090 GN=Edem3 PE=1 SV=2 |
|
| 1.85e-121 | 13 | 488 | 8 | 467 | Alpha-mannosidase I MNS4 OS=Arabidopsis thaliana OX=3702 GN=MNS4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | CS Position |
|---|---|---|
| 0.000330 | 0.999647 | CS pos: 29-30. Pr: 0.9661 |